Gene
KWMTBOMO13557
Pre Gene Modal
BGIBMGA010990
Annotation
PREDICTED:_DCN1-like_protein_5_[Amyelois_transitella]
Full name
Defective in cullin neddylation protein
Location in the cell
Cytoplasmic Reliability : 1.323 Nuclear Reliability : 1.814
Sequence
CDS
ATGCCTCGGAGCAGCAAACGAACGAGGAGCGGCGCACCTACAAGCGGAAACGATGTTCTGCCTGGTGCTACATCCATAGATGAACACCATCATCACAAAAGATCAAGAAATAGCAGTTTACGTCGACATTTGAAAGCAGAAGACACAAGTTTCAGTGTAAAGAAATGCTTATCATGGTTCAAAGAGTACACTTCGGTATCAGAACCGGATGTTTTAGGTCCTGAAGGAATGGAAAAGTTTTGTGCAGACCTAGGTGTAGACCCCGAAAATGTGGTGATGTTGGTTATAGCTTACAAAATGGGAGCGAAACAAATGGGATTTTTCACACAGGAAGAATGGTTAAAAGGCTTAACAGAGCTACAATGCGACAATGTACATAAATTACAAAATAAATTAGATTATTTACGGAATTTACTTAATGATCCACACATATTTAAGGCTATTTATCGATATTCATATGATTTTGCAAGGGACAAGGAGCAGCGCTCGCTGGACACGGAGGTGGCGGGCGCGCTGCTGGGCGTGCTGCTGCCGCGCTGGGCGCTGCGGGCGGCGCTGTGCGAGTTCATCGCGCGGGAGCGGCGGTACCGCGTCGTCAACCGCGACCAGTGGTGCAACATCCTCGAGTTCAGCCGCACCGTCGACGCGCACCTCGCCTCCTACGACGCCGACGGAGCCTGGCCAGTTATGTTAGACGAATTTGTGGAATGGTTAAGGGCGAACGACTCATCAATGCCCCCGGCCAGCTGA
Protein
MPRSSKRTRSGAPTSGNDVLPGATSIDEHHHHKRSRNSSLRRHLKAEDTSFSVKKCLSWFKEYTSVSEPDVLGPEGMEKFCADLGVDPENVVMLVIAYKMGAKQMGFFTQEEWLKGLTELQCDNVHKLQNKLDYLRNLLNDPHIFKAIYRYSYDFARDKEQRSLDTEVAGALLGVLLPRWALRAALCEFIARERRYRVVNRDQWCNILEFSRTVDAHLASYDADGAWPVMLDEFVEWLRANDSSMPPAS
Summary
Description
Neddylation of cullins play an essential role in the regulation of SCF-type complexes activity.
Uniprot
A0A3S2MAD4
A0A2H1W4G8
A0A212EXK0
A0A2W1BZM0
A0A2A4IY23
A0A194QE83
+ More
H9JN87 A0A194RJB2 A0A232ERV1 Q174N9 A0A1W7R566 A0A0T6AUK6 A0A2J7QQ16 A0A182QKG9 A0A1B6CRL4 A0A182UXE6 A0A182U2V5 A0A182XHW4 A0A182LEY0 Q7PGI3 A0A182HSU6 A0A182NRQ9 A0A1Y1L2Y5 A0A182IZB6 A0A0M9A206 A0A067RAQ9 A0A182P971 A0A0L7QZW2 A0A182LSK6 A0A1B0B3M5 A0A0K8TLX2 A0A2A3EL01 A0A088AKB9 A0A1B6LWI1 A0A1A9XQU1 A0A1A9UQS7 D3TQ09 A0A084VAQ8 A0A1B0EV37 A0A182VWS3 A0A2J7QQ06 A0A1I8JUP2 A0A0L0BQ66 A0A023EKV8 A0A1B6GP44 A0A0A9YQ07 A0A182FRP7 A0A146MFY7 E2AQ02 E2B8U4 K7IYC4 F5HKW8 A0A1B6JU56 A0A2M3ZBC9 A0A2M4ARP7 W5J8N0 A0A224XV86 A0A1Q3F4G1 E9IVF6 B0X114 N6TUV3 A0A0V0G689 U5EPX7 A0A195E2B8 A0A1I8MHY5 A0A0P4VT91 R4FLI4 T1DII9 A0A3L8DRM3 A0A2J7QQ29 E0VPE7 B4KXR6 A0A0C9PRC7 A0A161MNN8 A0A023FAV2 B4LFP7 A0A1Q3F4B0 A0A1I8JUM1 A0A151WGL2 F4WA62 A0A195C1H8 J3JZ67 A0A195AV80 A0A158NHK9 B4ITI8 D7EJW2 B3NIJ1 A0A1Q3F4Q1 A0A1I8PQ36 A0A1L8DSJ1 A0A195EUM3 A0A034WJJ8 A0A0A1WTQ6 B4GR13 B5DQM3 B3M4R8 A0A1I8PQA0 A0A1I8MHY7 B4IA75
H9JN87 A0A194RJB2 A0A232ERV1 Q174N9 A0A1W7R566 A0A0T6AUK6 A0A2J7QQ16 A0A182QKG9 A0A1B6CRL4 A0A182UXE6 A0A182U2V5 A0A182XHW4 A0A182LEY0 Q7PGI3 A0A182HSU6 A0A182NRQ9 A0A1Y1L2Y5 A0A182IZB6 A0A0M9A206 A0A067RAQ9 A0A182P971 A0A0L7QZW2 A0A182LSK6 A0A1B0B3M5 A0A0K8TLX2 A0A2A3EL01 A0A088AKB9 A0A1B6LWI1 A0A1A9XQU1 A0A1A9UQS7 D3TQ09 A0A084VAQ8 A0A1B0EV37 A0A182VWS3 A0A2J7QQ06 A0A1I8JUP2 A0A0L0BQ66 A0A023EKV8 A0A1B6GP44 A0A0A9YQ07 A0A182FRP7 A0A146MFY7 E2AQ02 E2B8U4 K7IYC4 F5HKW8 A0A1B6JU56 A0A2M3ZBC9 A0A2M4ARP7 W5J8N0 A0A224XV86 A0A1Q3F4G1 E9IVF6 B0X114 N6TUV3 A0A0V0G689 U5EPX7 A0A195E2B8 A0A1I8MHY5 A0A0P4VT91 R4FLI4 T1DII9 A0A3L8DRM3 A0A2J7QQ29 E0VPE7 B4KXR6 A0A0C9PRC7 A0A161MNN8 A0A023FAV2 B4LFP7 A0A1Q3F4B0 A0A1I8JUM1 A0A151WGL2 F4WA62 A0A195C1H8 J3JZ67 A0A195AV80 A0A158NHK9 B4ITI8 D7EJW2 B3NIJ1 A0A1Q3F4Q1 A0A1I8PQ36 A0A1L8DSJ1 A0A195EUM3 A0A034WJJ8 A0A0A1WTQ6 B4GR13 B5DQM3 B3M4R8 A0A1I8PQA0 A0A1I8MHY7 B4IA75
Pubmed
22118469
28756777
26354079
19121390
28648823
17510324
+ More
20966253 12364791 14747013 17210077 28004739 24845553 26369729 20353571 24438588 26108605 24945155 25401762 26823975 20798317 20075255 20920257 23761445 21282665 23537049 25315136 27129103 24330624 30249741 20566863 17994087 18057021 25474469 21719571 22516182 21347285 17550304 18362917 19820115 25348373 25830018 15632085 23185243
20966253 12364791 14747013 17210077 28004739 24845553 26369729 20353571 24438588 26108605 24945155 25401762 26823975 20798317 20075255 20920257 23761445 21282665 23537049 25315136 27129103 24330624 30249741 20566863 17994087 18057021 25474469 21719571 22516182 21347285 17550304 18362917 19820115 25348373 25830018 15632085 23185243
EMBL
RSAL01000002
RVE55033.1
RVE55086.1
ODYU01006282
SOQ47985.1
AGBW02011749
+ More
OWR46191.1 KZ150435 KZ149909 PZC70878.1 PZC78166.1 NWSH01004709 PCG64735.1 KQ459144 KPJ03729.1 BABH01033228 KQ460313 KPJ16031.1 NNAY01002553 OXU21067.1 CH477408 EAT41516.1 GEHC01001341 JAV46304.1 LJIG01022775 KRT78780.1 NEVH01012087 PNF30671.1 AXCN02000008 GEDC01021445 GEDC01003058 JAS15853.1 JAS34240.1 AAAB01008859 EAA44922.3 APCN01000361 GEZM01068157 GEZM01068156 JAV67131.1 KQ435759 KOX75675.1 KK852818 KDR15715.1 KQ414672 KOC64159.1 AXCM01001348 JXJN01007906 GDAI01002249 JAI15354.1 KZ288217 PBC32388.1 GEBQ01011952 JAT28025.1 CCAG010003291 EZ423511 ADD19787.1 ATLV01004137 KE524190 KFB35052.1 AJWK01014478 PNF30670.1 JRES01001542 KNC22151.1 GAPW01003791 JAC09807.1 GECZ01005646 JAS64123.1 GBHO01008512 GBRD01003397 GBRD01002010 GDHC01004657 JAG35092.1 JAG62424.1 JAQ13972.1 GDHC01001089 JAQ17540.1 GL441645 EFN64489.1 GL446384 EFN87884.1 EGK96870.1 GECU01005014 JAT02693.1 GGFM01005050 MBW25801.1 GGFK01010130 MBW43451.1 ADMH02001952 ETN60321.1 GFTR01004014 JAW12412.1 GFDL01012594 JAV22451.1 GL766234 EFZ15432.1 DS232250 EDS38406.1 APGK01053953 KB741239 KB632275 ENN72161.1 ERL91130.1 GECL01002611 JAP03513.1 GANO01003526 JAB56345.1 KQ979763 KYN19288.1 GDKW01001406 JAI55189.1 ACPB03003463 ACPB03003464 GAHY01001378 JAA76132.1 GALA01000906 JAA93946.1 QOIP01000005 RLU22952.1 PNF30673.1 DS235363 EEB15253.1 CH933809 EDW19773.1 GBYB01003813 JAG73580.1 GEMB01001029 JAS02118.1 GBBI01000659 JAC18053.1 CH940647 EDW70365.1 GFDL01012639 JAV22406.1 KQ983167 KYQ46947.1 GL888041 EGI68918.1 KQ978350 KYM94729.1 BT128548 AEE63505.1 KQ976737 KYM75962.1 ADTU01015917 CH891709 EDW99701.1 KQ971866 EFA12878.2 CH954178 EDV52487.1 GFDL01012494 JAV22551.1 GFDF01004638 JAV09446.1 KQ981975 KYN31597.1 GAKP01004658 GAKP01004657 JAC54295.1 GBXI01012060 JAD02232.1 CH479188 EDW40198.1 CH379069 EDY73444.1 CH902618 EDV39467.1 KPU78060.1 CH480826 EDW44188.1
OWR46191.1 KZ150435 KZ149909 PZC70878.1 PZC78166.1 NWSH01004709 PCG64735.1 KQ459144 KPJ03729.1 BABH01033228 KQ460313 KPJ16031.1 NNAY01002553 OXU21067.1 CH477408 EAT41516.1 GEHC01001341 JAV46304.1 LJIG01022775 KRT78780.1 NEVH01012087 PNF30671.1 AXCN02000008 GEDC01021445 GEDC01003058 JAS15853.1 JAS34240.1 AAAB01008859 EAA44922.3 APCN01000361 GEZM01068157 GEZM01068156 JAV67131.1 KQ435759 KOX75675.1 KK852818 KDR15715.1 KQ414672 KOC64159.1 AXCM01001348 JXJN01007906 GDAI01002249 JAI15354.1 KZ288217 PBC32388.1 GEBQ01011952 JAT28025.1 CCAG010003291 EZ423511 ADD19787.1 ATLV01004137 KE524190 KFB35052.1 AJWK01014478 PNF30670.1 JRES01001542 KNC22151.1 GAPW01003791 JAC09807.1 GECZ01005646 JAS64123.1 GBHO01008512 GBRD01003397 GBRD01002010 GDHC01004657 JAG35092.1 JAG62424.1 JAQ13972.1 GDHC01001089 JAQ17540.1 GL441645 EFN64489.1 GL446384 EFN87884.1 EGK96870.1 GECU01005014 JAT02693.1 GGFM01005050 MBW25801.1 GGFK01010130 MBW43451.1 ADMH02001952 ETN60321.1 GFTR01004014 JAW12412.1 GFDL01012594 JAV22451.1 GL766234 EFZ15432.1 DS232250 EDS38406.1 APGK01053953 KB741239 KB632275 ENN72161.1 ERL91130.1 GECL01002611 JAP03513.1 GANO01003526 JAB56345.1 KQ979763 KYN19288.1 GDKW01001406 JAI55189.1 ACPB03003463 ACPB03003464 GAHY01001378 JAA76132.1 GALA01000906 JAA93946.1 QOIP01000005 RLU22952.1 PNF30673.1 DS235363 EEB15253.1 CH933809 EDW19773.1 GBYB01003813 JAG73580.1 GEMB01001029 JAS02118.1 GBBI01000659 JAC18053.1 CH940647 EDW70365.1 GFDL01012639 JAV22406.1 KQ983167 KYQ46947.1 GL888041 EGI68918.1 KQ978350 KYM94729.1 BT128548 AEE63505.1 KQ976737 KYM75962.1 ADTU01015917 CH891709 EDW99701.1 KQ971866 EFA12878.2 CH954178 EDV52487.1 GFDL01012494 JAV22551.1 GFDF01004638 JAV09446.1 KQ981975 KYN31597.1 GAKP01004658 GAKP01004657 JAC54295.1 GBXI01012060 JAD02232.1 CH479188 EDW40198.1 CH379069 EDY73444.1 CH902618 EDV39467.1 KPU78060.1 CH480826 EDW44188.1
Proteomes
UP000283053
UP000007151
UP000218220
UP000053268
UP000005204
UP000053240
+ More
UP000215335 UP000008820 UP000235965 UP000075886 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075884 UP000075880 UP000053105 UP000027135 UP000075885 UP000053825 UP000075883 UP000092460 UP000242457 UP000005203 UP000092443 UP000078200 UP000092444 UP000030765 UP000092461 UP000075920 UP000075900 UP000037069 UP000069272 UP000000311 UP000008237 UP000002358 UP000000673 UP000002320 UP000019118 UP000030742 UP000078492 UP000095301 UP000015103 UP000279307 UP000009046 UP000009192 UP000008792 UP000075809 UP000007755 UP000078542 UP000078540 UP000005205 UP000002282 UP000007266 UP000008711 UP000095300 UP000078541 UP000008744 UP000001819 UP000007801 UP000001292
UP000215335 UP000008820 UP000235965 UP000075886 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075884 UP000075880 UP000053105 UP000027135 UP000075885 UP000053825 UP000075883 UP000092460 UP000242457 UP000005203 UP000092443 UP000078200 UP000092444 UP000030765 UP000092461 UP000075920 UP000075900 UP000037069 UP000069272 UP000000311 UP000008237 UP000002358 UP000000673 UP000002320 UP000019118 UP000030742 UP000078492 UP000095301 UP000015103 UP000279307 UP000009046 UP000009192 UP000008792 UP000075809 UP000007755 UP000078542 UP000078540 UP000005205 UP000002282 UP000007266 UP000008711 UP000095300 UP000078541 UP000008744 UP000001819 UP000007801 UP000001292
PRIDE
Pfam
PF03556 Cullin_binding
SUPFAM
SSF47473
SSF47473
ProteinModelPortal
A0A3S2MAD4
A0A2H1W4G8
A0A212EXK0
A0A2W1BZM0
A0A2A4IY23
A0A194QE83
+ More
H9JN87 A0A194RJB2 A0A232ERV1 Q174N9 A0A1W7R566 A0A0T6AUK6 A0A2J7QQ16 A0A182QKG9 A0A1B6CRL4 A0A182UXE6 A0A182U2V5 A0A182XHW4 A0A182LEY0 Q7PGI3 A0A182HSU6 A0A182NRQ9 A0A1Y1L2Y5 A0A182IZB6 A0A0M9A206 A0A067RAQ9 A0A182P971 A0A0L7QZW2 A0A182LSK6 A0A1B0B3M5 A0A0K8TLX2 A0A2A3EL01 A0A088AKB9 A0A1B6LWI1 A0A1A9XQU1 A0A1A9UQS7 D3TQ09 A0A084VAQ8 A0A1B0EV37 A0A182VWS3 A0A2J7QQ06 A0A1I8JUP2 A0A0L0BQ66 A0A023EKV8 A0A1B6GP44 A0A0A9YQ07 A0A182FRP7 A0A146MFY7 E2AQ02 E2B8U4 K7IYC4 F5HKW8 A0A1B6JU56 A0A2M3ZBC9 A0A2M4ARP7 W5J8N0 A0A224XV86 A0A1Q3F4G1 E9IVF6 B0X114 N6TUV3 A0A0V0G689 U5EPX7 A0A195E2B8 A0A1I8MHY5 A0A0P4VT91 R4FLI4 T1DII9 A0A3L8DRM3 A0A2J7QQ29 E0VPE7 B4KXR6 A0A0C9PRC7 A0A161MNN8 A0A023FAV2 B4LFP7 A0A1Q3F4B0 A0A1I8JUM1 A0A151WGL2 F4WA62 A0A195C1H8 J3JZ67 A0A195AV80 A0A158NHK9 B4ITI8 D7EJW2 B3NIJ1 A0A1Q3F4Q1 A0A1I8PQ36 A0A1L8DSJ1 A0A195EUM3 A0A034WJJ8 A0A0A1WTQ6 B4GR13 B5DQM3 B3M4R8 A0A1I8PQA0 A0A1I8MHY7 B4IA75
H9JN87 A0A194RJB2 A0A232ERV1 Q174N9 A0A1W7R566 A0A0T6AUK6 A0A2J7QQ16 A0A182QKG9 A0A1B6CRL4 A0A182UXE6 A0A182U2V5 A0A182XHW4 A0A182LEY0 Q7PGI3 A0A182HSU6 A0A182NRQ9 A0A1Y1L2Y5 A0A182IZB6 A0A0M9A206 A0A067RAQ9 A0A182P971 A0A0L7QZW2 A0A182LSK6 A0A1B0B3M5 A0A0K8TLX2 A0A2A3EL01 A0A088AKB9 A0A1B6LWI1 A0A1A9XQU1 A0A1A9UQS7 D3TQ09 A0A084VAQ8 A0A1B0EV37 A0A182VWS3 A0A2J7QQ06 A0A1I8JUP2 A0A0L0BQ66 A0A023EKV8 A0A1B6GP44 A0A0A9YQ07 A0A182FRP7 A0A146MFY7 E2AQ02 E2B8U4 K7IYC4 F5HKW8 A0A1B6JU56 A0A2M3ZBC9 A0A2M4ARP7 W5J8N0 A0A224XV86 A0A1Q3F4G1 E9IVF6 B0X114 N6TUV3 A0A0V0G689 U5EPX7 A0A195E2B8 A0A1I8MHY5 A0A0P4VT91 R4FLI4 T1DII9 A0A3L8DRM3 A0A2J7QQ29 E0VPE7 B4KXR6 A0A0C9PRC7 A0A161MNN8 A0A023FAV2 B4LFP7 A0A1Q3F4B0 A0A1I8JUM1 A0A151WGL2 F4WA62 A0A195C1H8 J3JZ67 A0A195AV80 A0A158NHK9 B4ITI8 D7EJW2 B3NIJ1 A0A1Q3F4Q1 A0A1I8PQ36 A0A1L8DSJ1 A0A195EUM3 A0A034WJJ8 A0A0A1WTQ6 B4GR13 B5DQM3 B3M4R8 A0A1I8PQA0 A0A1I8MHY7 B4IA75
PDB
5V89
E-value=2.06056e-65,
Score=630
Ontologies
GO
PANTHER
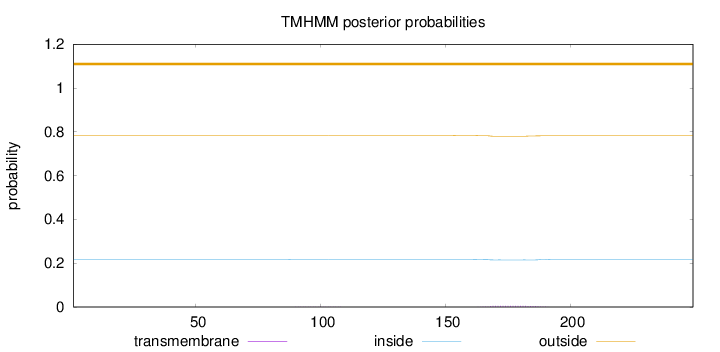
Topology
Length:
249
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11514
Exp number, first 60 AAs:
0
Total prob of N-in:
0.21779
outside
1 - 249
Population Genetic Test Statistics
Pi
156.860604
Theta
148.188147
Tajima's D
0.234852
CLR
0.496177
CSRT
0.428378581070946
Interpretation
Uncertain
