Gene
KWMTBOMO13553
Pre Gene Modal
BGIBMGA010976
Annotation
carboxyl/cholinesterase_1_precursor_[Bombyx_mori]
Full name
Carboxylic ester hydrolase
Location in the cell
PlasmaMembrane Reliability : 1.145
Sequence
CDS
ATGTTACTGTTTTATTTAATGTTTGGATTGTTGAGTGGTGTGGAATTGGTGAAAGTTAAAATAACCGAAGGATGGTTGAGAGGTGAATTGTTGGAGGCTTCAACGGGAGACAAGAAATATTACAGTTTCAAAGGAATACCTTATGCGGCGCCACCTCTTGGAAATTTAAGATTTGTGGCTCCTCAACCGCCATTATCCTGGAAAAACGTCAGGGACGCCACACGTCATGGTCCGAAATGCATTCAGACGGAGACTTTGTCGACCAAGTTGGTCCCCGGGAGTGAAGACTGTCTTTTTATCAACGTATACAGTCCAGACCTTAAACCCAAAGAGCAACTACCGGTTATGTTTTTCATATACGGCGGTGGATATAAATCAGGCTCTGGTGATGTCGACGATTATGGACCAGACTTTTTAATAAGATATGACGTAGTCGTCGTGACATTCAACTATAGGCTCGGCGCTTTGGGGTTTTTATGTACAGAAACTAAAGAAGTTCCAGGTAATGCTGGAATGAAAGACCAGGTTGCTGCACTGCGTTGGGTAAAAAGTAACATTGCCCATTTTGGTGGGAACCCAGATCAAGTCACGATTTTTGGACAAAGCGCTGGTGGTGCTTCTGTGGCTTTGCATACTTTGTCTCCCTTATCCAAAGGCTTATTTAAGCGGGCTATTGTAATGAGTGATTCATTCTATAACGACCCATTAAGACCGTTTGAATCTAAAATGAGGGCTTTGGTGTTAGGGAAACGATTGGGCTTAGACACGGCTAATACCACTGAACTTTTGGATTATCTACGGTGTGTGCCCGCTGAAAAGCTATTCATCCAACATCCTAATGTAATAATAACGGAGAGCATCGCTCAAAGTGTTATAAAATTATTCTATTTCACCCCAGTCGTGGAGAAAGATTTCGGTGATGATCATTTTTTGACTGAAGATCCCGAGGTGCTACTCTCTCAAGGTAAAATCCAACAAGACGTCGACTTAATAGTTGGGCACACGAATATAGAAGGTATTTACAATGTGGCTCAGCTTCAGGATAGAGTCCTGAAATATTATCCTTTATACCAGGAACTTTTGGTGCCTCGTAAAATAAGCAACATAAGAGCCCCAACAAAAGTTCTAGAGTTATCTCGTTTGATCTACAGATACTACTTCGGCGACAGACCAATCAATTTGGATACTATCAAAGAATTCGTTTACTACTACACCGACGCGAACTATGCTTATGATGTTTATAGATTTATAAACAAGTTTCCTCAAAGTGCTCACAAGAATCGGTTCTATTACATCTTTTCACCGTATACAGAGAGGAATTATTATTCAAGACTAGGCGCTGAGCGTTATGGCCTATACGGGTCAGCTCATAGAGATGATCTGATGTACTTGTTCGACGGAAAACGATTTAATTTGACTTTAGATAAGAATAGTACGTCGTTCAAGATTATAAATAATTTCTGTGCCTTGTTGACGAACTTCGCTAAATTTGGGAATCCAACACCAGATAATTCTGGAAACGTGATTTGGCCAAAATTTGATAAAGAACAGCGATACCTGAACATTGGAAAAGAGCTTACAGCTAGAAGAATTATGTTTCAGCAGAGTTCTCTTTCATTTTGGAAGAATATTCTAGAGCGAGCCGAACTTGAATTTTAA
Protein
MLLFYLMFGLLSGVELVKVKITEGWLRGELLEASTGDKKYYSFKGIPYAAPPLGNLRFVAPQPPLSWKNVRDATRHGPKCIQTETLSTKLVPGSEDCLFINVYSPDLKPKEQLPVMFFIYGGGYKSGSGDVDDYGPDFLIRYDVVVVTFNYRLGALGFLCTETKEVPGNAGMKDQVAALRWVKSNIAHFGGNPDQVTIFGQSAGGASVALHTLSPLSKGLFKRAIVMSDSFYNDPLRPFESKMRALVLGKRLGLDTANTTELLDYLRCVPAEKLFIQHPNVIITESIAQSVIKLFYFTPVVEKDFGDDHFLTEDPEVLLSQGKIQQDVDLIVGHTNIEGIYNVAQLQDRVLKYYPLYQELLVPRKISNIRAPTKVLELSRLIYRYYFGDRPINLDTIKEFVYYYTDANYAYDVYRFINKFPQSAHKNRFYYIFSPYTERNYYSRLGAERYGLYGSAHRDDLMYLFDGKRFNLTLDKNSTSFKIINNFCALLTNFAKFGNPTPDNSGNVIWPKFDKEQRYLNIGKELTARRIMFQQSSLSFWKNILERAELEF
Summary
Similarity
Belongs to the type-B carboxylesterase/lipase family.
Feature
chain Carboxylic ester hydrolase
Uniprot
D2KTU3
H9JN73
A0A2S0D881
H9JN84
A9LS22
A0A076FRM1
+ More
A0A2W1BKQ4 A0A2A4JEW3 A0A2U3T8L5 A0A0K8TUL7 A0A2U3T8J6 A0A2A4JFQ0 A0A0K8TVU9 A0A194REM3 A0A291P123 A0A1U9X1T3 J7HWI3 A0A0L7KNG8 A0A194QDV9 A0A1J0M1C9 A0A2W1BR44 A0A2H1VCP7 F1C4B8 D3GDL4 A0A0K0XRL4 A0A2U3T8L4 A0A3S2MAW7 A0A0Y0B016 A0A2U3T8L2 A0A0L7KL89 A0A109QN31 U5KKB6 D5G3F2 A3QR02 A0A3S2TTT0 A0A2W1BKK3 A0A1U9X1T8 A0A2U3T8L3 A0A2H1WNK8 A0A346TPC2 A0A1U9X1X0 A0A291P0Q3 A0A1U9X1U2 A0A2H1VCP1 A0A0K8TVW5 A0A0F7QEB1 A0A0Y0BIG3 A0A0K8TV87 A0A0B4RY15 A0A194RE38 A0A291P0X6 D5G3F1 D5G3F0 A0A2W1BUX0 A0A2H1VCU4 A0A291P0R0 A0A291P0V0 A0A194PSZ9 A0A2W1BGL8 A0A2A4K7Y3 A0A2W1BMW3 A0A2I6RJN9 A0A2A4JIZ5 D5G3F3 A0A088QEL8 A0A1L8D6M2 A0A194RDX2 D3GDM1 H9JN83 A0A2W1BSV8 A0A1U9X1S5 G1C2I6 A0A291P0Y3 F8V329 A0A1L8D6J3 A0A0C5C1M2 A0A194RJI0 A0A2H1WNU4 A0A2A4JJM6 A0A076FT12 A0A2H1V381 A0A194PRF0 A0A194PT59 A0A194PSA6 A0A291P117 Q1W2E3 A0A291P0V7 A0A194PSE5 A0A2H1WFF3 G3CM70 A0A291P0U0 D5G3F4 A0A0Y0BA98 A0A0K8TVA5 A0A194RDD6 A0A2W1C223 A0A2S0D942 A0A1L8D6J2
A0A2W1BKQ4 A0A2A4JEW3 A0A2U3T8L5 A0A0K8TUL7 A0A2U3T8J6 A0A2A4JFQ0 A0A0K8TVU9 A0A194REM3 A0A291P123 A0A1U9X1T3 J7HWI3 A0A0L7KNG8 A0A194QDV9 A0A1J0M1C9 A0A2W1BR44 A0A2H1VCP7 F1C4B8 D3GDL4 A0A0K0XRL4 A0A2U3T8L4 A0A3S2MAW7 A0A0Y0B016 A0A2U3T8L2 A0A0L7KL89 A0A109QN31 U5KKB6 D5G3F2 A3QR02 A0A3S2TTT0 A0A2W1BKK3 A0A1U9X1T8 A0A2U3T8L3 A0A2H1WNK8 A0A346TPC2 A0A1U9X1X0 A0A291P0Q3 A0A1U9X1U2 A0A2H1VCP1 A0A0K8TVW5 A0A0F7QEB1 A0A0Y0BIG3 A0A0K8TV87 A0A0B4RY15 A0A194RE38 A0A291P0X6 D5G3F1 D5G3F0 A0A2W1BUX0 A0A2H1VCU4 A0A291P0R0 A0A291P0V0 A0A194PSZ9 A0A2W1BGL8 A0A2A4K7Y3 A0A2W1BMW3 A0A2I6RJN9 A0A2A4JIZ5 D5G3F3 A0A088QEL8 A0A1L8D6M2 A0A194RDX2 D3GDM1 H9JN83 A0A2W1BSV8 A0A1U9X1S5 G1C2I6 A0A291P0Y3 F8V329 A0A1L8D6J3 A0A0C5C1M2 A0A194RJI0 A0A2H1WNU4 A0A2A4JJM6 A0A076FT12 A0A2H1V381 A0A194PRF0 A0A194PT59 A0A194PSA6 A0A291P117 Q1W2E3 A0A291P0V7 A0A194PSE5 A0A2H1WFF3 G3CM70 A0A291P0U0 D5G3F4 A0A0Y0BA98 A0A0K8TVA5 A0A194RDD6 A0A2W1C223 A0A2S0D942 A0A1L8D6J2
EC Number
3.1.1.-
Pubmed
EMBL
AB299280
BAI66477.1
BABH01033215
KX015872
ARM65401.1
BABH01033219
+ More
EU255790 ABX46627.1 KF960778 AII21980.1 KZ150061 PZC74214.1 NWSH01001642 PCG70597.1 KX015873 ARM65402.1 GCVX01000041 JAI18189.1 KX015845 ARM65374.1 PCG70596.1 GCVX01000040 JAI18190.1 KQ460313 KPJ16032.1 MF687624 ATJ44550.1 KY021826 AQY62715.1 JX489387 GEYN01000055 AFQ20792.1 JAV02074.1 JTDY01007971 KOB64827.1 KQ459144 KPJ03728.1 KU375427 APD13869.1 PZC74213.1 ODYU01001848 SOQ38619.1 HQ199328 ADX30519.1 FJ652446 ACV60230.1 KP938873 AKS40354.1 KX015871 ARM65400.1 RSAL01000002 RVE55036.1 RVE55089.1 KU215369 AMB19663.1 KX015869 ARM65398.1 JTDY01009253 KOB63826.1 KU215373 AMB19667.1 JX866739 AGS49143.1 FJ997316 ADF43481.1 DQ401086 ABD62772.1 RVE55035.1 RVE55088.1 PZC74215.1 KY021833 AQY62722.1 KX015870 ARM65399.1 ODYU01009917 SOQ54655.1 MF589742 AXU41412.1 KY021861 AQY62750.1 MF687565 ATJ44491.1 KY021834 AQY62723.1 SOQ38618.1 GCVX01000025 JAI18205.1 LC017767 BAR64781.1 KU215366 AMB19660.1 GCVX01000032 JAI18198.1 KJ023215 AIY69043.1 KPJ16098.1 MF687625 ATJ44551.1 FJ997315 ADF43480.1 FJ997314 ADF43479.1 KZ149909 PZC78161.1 SOQ38621.1 MF687557 ATJ44483.1 MF687563 ATJ44489.1 KQ459595 KPI95889.1 PZC74212.1 NWSH01000066 PCG80014.1 PZC74216.1 KY304476 AUN86407.1 NWSH01001228 PCG72037.1 FJ997317 ADF43482.1 KM008609 AIN76405.1 GEYN01000026 JAV02103.1 KPJ16038.1 FJ652453 ACV60237.1 BABH01033216 PZC78162.1 KY021821 AQY62710.1 JF728805 AEJ38207.1 MF687626 ATJ44552.1 JN019808 AEJ18172.1 GEYN01000056 JAV02073.1 KP001161 AJN91206.1 KPJ16096.1 ODYU01009893 SOQ54612.1 PCG72029.1 KF960782 AII21984.1 ODYU01000269 SOQ34744.1 KPI95887.1 KPI95949.1 KPI95848.1 MF687633 ATJ44559.1 DQ445461 ABE01157.2 MF687564 ATJ44490.1 KPI95888.1 ODYU01008252 SOQ51676.1 HQ122616 AEL33699.1 MF687553 ATJ44479.1 FJ997318 ADF43483.1 KU215371 AMB19665.1 GCVX01000034 JAI18196.1 KQ460323 KPJ15853.1 PZC78163.1 KX015867 ARM65396.1 GEYN01000057 JAV02072.1
EU255790 ABX46627.1 KF960778 AII21980.1 KZ150061 PZC74214.1 NWSH01001642 PCG70597.1 KX015873 ARM65402.1 GCVX01000041 JAI18189.1 KX015845 ARM65374.1 PCG70596.1 GCVX01000040 JAI18190.1 KQ460313 KPJ16032.1 MF687624 ATJ44550.1 KY021826 AQY62715.1 JX489387 GEYN01000055 AFQ20792.1 JAV02074.1 JTDY01007971 KOB64827.1 KQ459144 KPJ03728.1 KU375427 APD13869.1 PZC74213.1 ODYU01001848 SOQ38619.1 HQ199328 ADX30519.1 FJ652446 ACV60230.1 KP938873 AKS40354.1 KX015871 ARM65400.1 RSAL01000002 RVE55036.1 RVE55089.1 KU215369 AMB19663.1 KX015869 ARM65398.1 JTDY01009253 KOB63826.1 KU215373 AMB19667.1 JX866739 AGS49143.1 FJ997316 ADF43481.1 DQ401086 ABD62772.1 RVE55035.1 RVE55088.1 PZC74215.1 KY021833 AQY62722.1 KX015870 ARM65399.1 ODYU01009917 SOQ54655.1 MF589742 AXU41412.1 KY021861 AQY62750.1 MF687565 ATJ44491.1 KY021834 AQY62723.1 SOQ38618.1 GCVX01000025 JAI18205.1 LC017767 BAR64781.1 KU215366 AMB19660.1 GCVX01000032 JAI18198.1 KJ023215 AIY69043.1 KPJ16098.1 MF687625 ATJ44551.1 FJ997315 ADF43480.1 FJ997314 ADF43479.1 KZ149909 PZC78161.1 SOQ38621.1 MF687557 ATJ44483.1 MF687563 ATJ44489.1 KQ459595 KPI95889.1 PZC74212.1 NWSH01000066 PCG80014.1 PZC74216.1 KY304476 AUN86407.1 NWSH01001228 PCG72037.1 FJ997317 ADF43482.1 KM008609 AIN76405.1 GEYN01000026 JAV02103.1 KPJ16038.1 FJ652453 ACV60237.1 BABH01033216 PZC78162.1 KY021821 AQY62710.1 JF728805 AEJ38207.1 MF687626 ATJ44552.1 JN019808 AEJ18172.1 GEYN01000056 JAV02073.1 KP001161 AJN91206.1 KPJ16096.1 ODYU01009893 SOQ54612.1 PCG72029.1 KF960782 AII21984.1 ODYU01000269 SOQ34744.1 KPI95887.1 KPI95949.1 KPI95848.1 MF687633 ATJ44559.1 DQ445461 ABE01157.2 MF687564 ATJ44490.1 KPI95888.1 ODYU01008252 SOQ51676.1 HQ122616 AEL33699.1 MF687553 ATJ44479.1 FJ997318 ADF43483.1 KU215371 AMB19665.1 GCVX01000034 JAI18196.1 KQ460323 KPJ15853.1 PZC78163.1 KX015867 ARM65396.1 GEYN01000057 JAV02072.1
Proteomes
Pfam
PF00135 COesterase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
D2KTU3
H9JN73
A0A2S0D881
H9JN84
A9LS22
A0A076FRM1
+ More
A0A2W1BKQ4 A0A2A4JEW3 A0A2U3T8L5 A0A0K8TUL7 A0A2U3T8J6 A0A2A4JFQ0 A0A0K8TVU9 A0A194REM3 A0A291P123 A0A1U9X1T3 J7HWI3 A0A0L7KNG8 A0A194QDV9 A0A1J0M1C9 A0A2W1BR44 A0A2H1VCP7 F1C4B8 D3GDL4 A0A0K0XRL4 A0A2U3T8L4 A0A3S2MAW7 A0A0Y0B016 A0A2U3T8L2 A0A0L7KL89 A0A109QN31 U5KKB6 D5G3F2 A3QR02 A0A3S2TTT0 A0A2W1BKK3 A0A1U9X1T8 A0A2U3T8L3 A0A2H1WNK8 A0A346TPC2 A0A1U9X1X0 A0A291P0Q3 A0A1U9X1U2 A0A2H1VCP1 A0A0K8TVW5 A0A0F7QEB1 A0A0Y0BIG3 A0A0K8TV87 A0A0B4RY15 A0A194RE38 A0A291P0X6 D5G3F1 D5G3F0 A0A2W1BUX0 A0A2H1VCU4 A0A291P0R0 A0A291P0V0 A0A194PSZ9 A0A2W1BGL8 A0A2A4K7Y3 A0A2W1BMW3 A0A2I6RJN9 A0A2A4JIZ5 D5G3F3 A0A088QEL8 A0A1L8D6M2 A0A194RDX2 D3GDM1 H9JN83 A0A2W1BSV8 A0A1U9X1S5 G1C2I6 A0A291P0Y3 F8V329 A0A1L8D6J3 A0A0C5C1M2 A0A194RJI0 A0A2H1WNU4 A0A2A4JJM6 A0A076FT12 A0A2H1V381 A0A194PRF0 A0A194PT59 A0A194PSA6 A0A291P117 Q1W2E3 A0A291P0V7 A0A194PSE5 A0A2H1WFF3 G3CM70 A0A291P0U0 D5G3F4 A0A0Y0BA98 A0A0K8TVA5 A0A194RDD6 A0A2W1C223 A0A2S0D942 A0A1L8D6J2
A0A2W1BKQ4 A0A2A4JEW3 A0A2U3T8L5 A0A0K8TUL7 A0A2U3T8J6 A0A2A4JFQ0 A0A0K8TVU9 A0A194REM3 A0A291P123 A0A1U9X1T3 J7HWI3 A0A0L7KNG8 A0A194QDV9 A0A1J0M1C9 A0A2W1BR44 A0A2H1VCP7 F1C4B8 D3GDL4 A0A0K0XRL4 A0A2U3T8L4 A0A3S2MAW7 A0A0Y0B016 A0A2U3T8L2 A0A0L7KL89 A0A109QN31 U5KKB6 D5G3F2 A3QR02 A0A3S2TTT0 A0A2W1BKK3 A0A1U9X1T8 A0A2U3T8L3 A0A2H1WNK8 A0A346TPC2 A0A1U9X1X0 A0A291P0Q3 A0A1U9X1U2 A0A2H1VCP1 A0A0K8TVW5 A0A0F7QEB1 A0A0Y0BIG3 A0A0K8TV87 A0A0B4RY15 A0A194RE38 A0A291P0X6 D5G3F1 D5G3F0 A0A2W1BUX0 A0A2H1VCU4 A0A291P0R0 A0A291P0V0 A0A194PSZ9 A0A2W1BGL8 A0A2A4K7Y3 A0A2W1BMW3 A0A2I6RJN9 A0A2A4JIZ5 D5G3F3 A0A088QEL8 A0A1L8D6M2 A0A194RDX2 D3GDM1 H9JN83 A0A2W1BSV8 A0A1U9X1S5 G1C2I6 A0A291P0Y3 F8V329 A0A1L8D6J3 A0A0C5C1M2 A0A194RJI0 A0A2H1WNU4 A0A2A4JJM6 A0A076FT12 A0A2H1V381 A0A194PRF0 A0A194PT59 A0A194PSA6 A0A291P117 Q1W2E3 A0A291P0V7 A0A194PSE5 A0A2H1WFF3 G3CM70 A0A291P0U0 D5G3F4 A0A0Y0BA98 A0A0K8TVA5 A0A194RDD6 A0A2W1C223 A0A2S0D942 A0A1L8D6J2
PDB
5TYP
E-value=1.88512e-70,
Score=677
Ontologies
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.848878
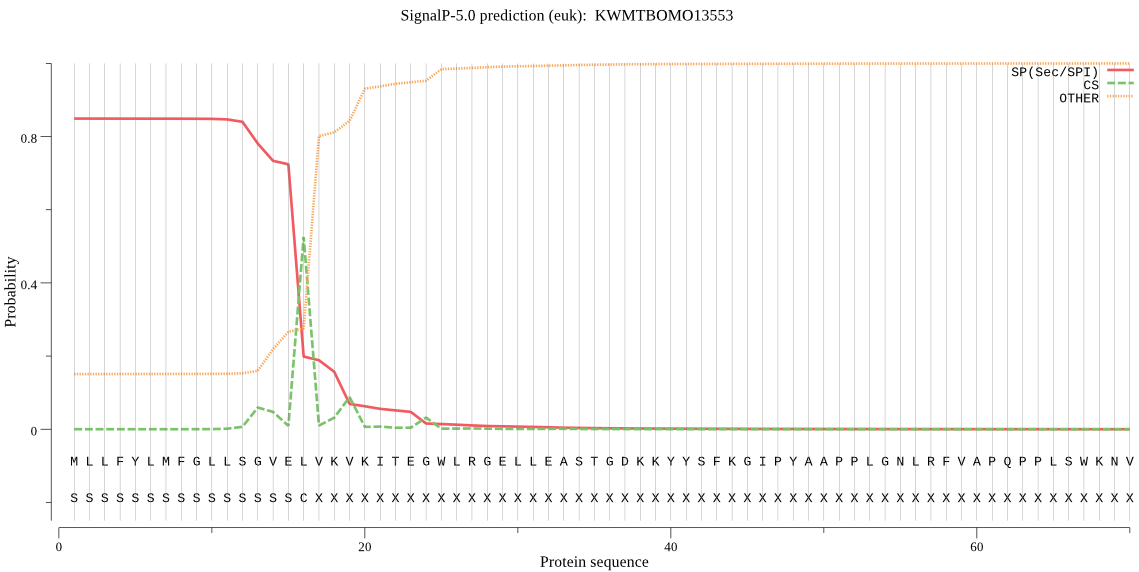
Length:
552
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01167
Exp number, first 60 AAs:
0.00932
Total prob of N-in:
0.00054
outside
1 - 552
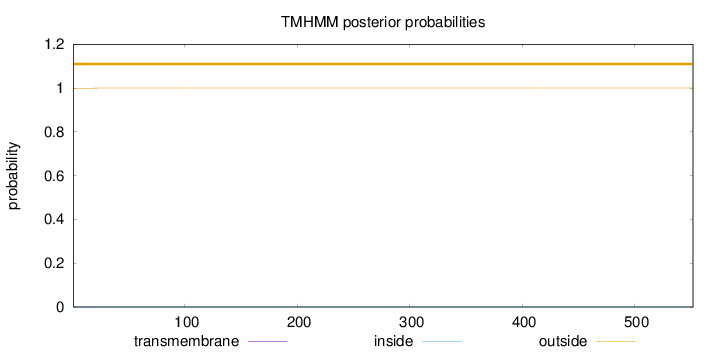
Population Genetic Test Statistics
Pi
284.428523
Theta
210.320559
Tajima's D
1.263721
CLR
0.182064
CSRT
0.728663566821659
Interpretation
Uncertain
