Gene
KWMTBOMO13530 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001307
Annotation
superoxide_dismutase_[Cu-Zn]_[Bombyx_mori]
Full name
Superoxide dismutase [Cu-Zn]
Alternative Name
Superoxide dismutase 1
Location in the cell
Cytoplasmic Reliability : 3.742
Sequence
CDS
ATGCCCGCCAAAGCAGTTTGCGTACTTCGTGGTGATGTTAGCGGAACTGTTTTCTTCGATCAGCAGGATGAAAAGTCACCTGTTGTTGTTTCTGGAGAGGTCCAGGGCTTGACTAAGGGTAAACACGGTTTCCATGTGCACGAATTTGGTGACAACACAAACGGTTGCACATCAGCTGGAGCTCATTTCAACCCTGAAAAACAAGATCATGGTGGTCCCAGTTCTGCTGTACGCCATGTCGGCGACCTCGGTAACATTGAGGCAATTGAAGACTCTGGAGTCACTAAAGTATCAATCCAAGATTCCCAGATCTCTCTTCATGGACCTAACAGCATCATTGGTCGCACTTTAGTTGTCCATGCTGACCCTGATGACTTGGGACTCGGTGGCCATGAGCTAAGTAAGACCACTGGTAATGCCGGTGGCCGTATTGCTTGTGGAGTCATTGGCTTGGCCAAGATTTAA
Protein
MPAKAVCVLRGDVSGTVFFDQQDEKSPVVVSGEVQGLTKGKHGFHVHEFGDNTNGCTSAGAHFNPEKQDHGGPSSAVRHVGDLGNIEAIEDSGVTKVSIQDSQISLHGPNSIIGRTLVVHADPDDLGLGGHELSKTTGNAGGRIACGVIGLAKI
Summary
Description
Destroys radicals which are normally produced within the cells and which are toxic to biological systems.
Catalytic Activity
2 H(+) + 2 superoxide = H2O2 + O2
Cofactor
Cu cation
Zn(2+)
Zn(2+)
Subunit
Homodimer.
Similarity
Belongs to the Cu-Zn superoxide dismutase family.
Keywords
3D-structure
Antioxidant
Complete proteome
Copper
Cytoplasm
Direct protein sequencing
Disulfide bond
Metal-binding
Oxidoreductase
Reference proteome
Zinc
Feature
chain Superoxide dismutase [Cu-Zn]
Uniprot
Q5TLU4
A0JKF3
P82205
A7BI63
A0A170QHN4
A0A1B1RSA9
+ More
A0A212FDT2 S4PG22 A0A2Z4N444 A0A194PR57 I4DJ15 A0A3S5HJH2 G4XH81 A0A2A4JHM4 A0A194RFH4 A0A2S0RQR7 C3RSN1 A0A2H1V9V5 H9BEW2 A0A0L7LLQ0 A0A2W1BUB5 Q60FS5 A0A1I8N7L1 Q6SCL6 A0A0U2SX80 A0A0S2A460 A0A1L8E4S1 A0A1B0CUF9 A0A1L8EFB3 A0A0S2A496 A0A1I8Q6I1 A0A1A9XY61 A0A1B0BND7 E0W322 A0A1W4W7H5 A0A1A9UFW7 A0A076G467 A0A023EH27 U5EW44 A0A109WQA1 A0A1A9ZKK9 A0A2R4LGJ8 D2A2T2 Q1HRN3 A0A385HVY4 A0A1Y1MBB2 R4V538 D3TRM9 A0A0C9RW85 M1JNK3 F4MI33 A0A0L0BQ22 A0A0N7EJN0 A0A0K8TQF3 A0A3G9D4Q3 W8BWU5 P28755 A0A2J7PGY8 A0A1A9W501 A0A1Q3FJF2 A0A2P1H680 Q6W5R8 A0A0A1WUB5 A0A067R0S2 A0A154P938 E2A8Y6 A6YPT7 R4WNL0 A0A023FAY3 A0A2Z4G643 A0A0V0G2Z1 Q7YXM6 A0A088A933 A0A1B6KJY5 V9IJ95 H6UWL0 A0A0P4VTN9 T1HRT6 A0A195AZY4 A0A158NMI6 A0A026WG66 B0X9L3 A0A1B6CIU5 P41973 B4IY00 Q103C4 Q07182 A0A1S3D1W5 A0A0M5J141 A0A069DPF3 A0A226DYR0 E3VSS4 C5NS97 D2Y3W7 B8YNX4 A0A0L7R2N7 G9C5D1 G9D320 A0A182GVF6 A0A2C9GUA8
A0A212FDT2 S4PG22 A0A2Z4N444 A0A194PR57 I4DJ15 A0A3S5HJH2 G4XH81 A0A2A4JHM4 A0A194RFH4 A0A2S0RQR7 C3RSN1 A0A2H1V9V5 H9BEW2 A0A0L7LLQ0 A0A2W1BUB5 Q60FS5 A0A1I8N7L1 Q6SCL6 A0A0U2SX80 A0A0S2A460 A0A1L8E4S1 A0A1B0CUF9 A0A1L8EFB3 A0A0S2A496 A0A1I8Q6I1 A0A1A9XY61 A0A1B0BND7 E0W322 A0A1W4W7H5 A0A1A9UFW7 A0A076G467 A0A023EH27 U5EW44 A0A109WQA1 A0A1A9ZKK9 A0A2R4LGJ8 D2A2T2 Q1HRN3 A0A385HVY4 A0A1Y1MBB2 R4V538 D3TRM9 A0A0C9RW85 M1JNK3 F4MI33 A0A0L0BQ22 A0A0N7EJN0 A0A0K8TQF3 A0A3G9D4Q3 W8BWU5 P28755 A0A2J7PGY8 A0A1A9W501 A0A1Q3FJF2 A0A2P1H680 Q6W5R8 A0A0A1WUB5 A0A067R0S2 A0A154P938 E2A8Y6 A6YPT7 R4WNL0 A0A023FAY3 A0A2Z4G643 A0A0V0G2Z1 Q7YXM6 A0A088A933 A0A1B6KJY5 V9IJ95 H6UWL0 A0A0P4VTN9 T1HRT6 A0A195AZY4 A0A158NMI6 A0A026WG66 B0X9L3 A0A1B6CIU5 P41973 B4IY00 Q103C4 Q07182 A0A1S3D1W5 A0A0M5J141 A0A069DPF3 A0A226DYR0 E3VSS4 C5NS97 D2Y3W7 B8YNX4 A0A0L7R2N7 G9C5D1 G9D320 A0A182GVF6 A0A2C9GUA8
EC Number
1.15.1.1
Pubmed
15784978
11280994
22118469
23622113
26354079
22651552
+ More
26227816 28756777 25315136 20566863 25042129 24945155 26483478 18362917 19820115 17204158 17510324 28004739 20353571 25348373 21569254 26108605 26369729 26760975 24495485 1342926 29527172 14993600 25830018 24845553 20798317 18207082 23691247 25474469 29889877 11932240 16139867 27129103 21347285 24508170 30249741 7926818 17994087 16730205 1343773 26334808 21835214 21937434
26227816 28756777 25315136 20566863 25042129 24945155 26483478 18362917 19820115 17204158 17510324 28004739 20353571 25348373 21569254 26108605 26369729 26760975 24495485 1342926 29527172 14993600 25830018 24845553 20798317 18207082 23691247 25474469 29889877 11932240 16139867 27129103 21347285 24508170 30249741 7926818 17994087 16730205 1343773 26334808 21835214 21937434
EMBL
AB179561
LC377907
BAD69805.2
BBD52302.1
EF077625
AM410997
+ More
ABK60176.1 CAL69462.1 AY461705 AAR97568.1 AB290453 BAF73670.1 KT984953 AMY63172.1 KU549181 ANU04687.1 AGBW02008999 OWR51892.1 GAIX01003807 JAA88753.1 MG431817 AWX63640.1 KQ459595 KPI95936.1 AK401283 BAM17905.1 MH374163 AZK16057.1 JF811436 AEP43785.1 NWSH01001445 PCG71218.1 KQ460313 KPJ16050.1 MF797881 RSAL01000069 AWA45435.1 RVE49227.1 EU263634 ABX11259.1 ODYU01001173 SOQ37014.1 JQ009331 AFC98366.1 JTDY01000644 KOB76355.1 KZ149964 PZC76236.1 AB189024 BAD52256.1 AY460107 KA644700 AAR23787.1 AFP59329.1 KT270580 ALR99794.1 KT002138 KT002143 ALN12440.1 ALN12445.1 GFDF01000552 JAV13532.1 AJWK01029153 GFDG01001520 JAV17279.1 KT002146 ALN12448.1 JXJN01017347 DS235881 EEB20028.1 KF527568 AII26027.1 JXUM01073365 GAPW01005443 GAPW01005442 GAPW01005441 GAPW01005440 KQ562770 JAC08156.1 KXJ75156.1 GANO01001597 JAB58274.1 KP642166 ALM23457.1 MH006694 AVW80115.1 KP342269 KQ971339 AKH80544.1 EFA02790.1 DQ440061 CH477769 CH477190 ABF18094.1 EAT36424.1 EAT48702.1 MG601765 AXY54993.1 GEZM01037908 JAV81850.1 KC741174 AGM32998.1 CCAG010001380 EZ424081 ADD20357.1 GBYB01012041 JAG81808.1 JX263431 GAKP01008857 AGE89777.1 JAC50095.1 EZ617714 ADH94607.1 JRES01001549 KNC22106.1 KR072549 ALF39391.1 GDAI01001488 JAI16115.1 FX985484 BAX90092.1 GAMC01002843 JAC03713.1 M76975 NEVH01025158 PNF15606.1 GFDL01007346 JAV27699.1 MF776705 AVN67038.1 AY309973 AAQ81639.1 GBXI01012274 JAD02018.1 KK853022 KDR12362.1 KQ434829 KZC07670.1 GL437703 EFN70085.1 EF639098 GEMB01003510 ABR27983.1 JAR99707.1 AK417231 BAN20446.1 GBBI01000075 JAC18637.1 MG983778 AWV96613.1 GECL01003765 JAP02359.1 AY329355 AAP93581.1 GEBQ01028209 JAT11768.1 JR050197 KM244765 AEY61198.1 AIU41504.1 JN700517 KZ288318 AEZ57109.1 PBC28279.1 GDKW01000598 JAI55997.1 ACPB03006759 KQ976697 KYM77514.1 ADTU01020601 KK107238 QOIP01000007 EZA54646.1 RLU20399.1 DS232541 EDS43182.1 GEDC01023958 JAS13340.1 L13281 CH964101 CH916366 EDV97543.1 DQ096569 AAZ79896.1 X61687 CP012525 ALC44136.1 GBGD01003323 JAC85566.1 LNIX01000009 OXA49934.1 HQ230310 ADO20320.1 AB475105 BAH83704.1 GU300109 ADB54843.2 FJ554532 HQ326578 HQ326579 HQ326580 HQ326581 HQ326582 HQ326583 ACL80663.2 ADO79626.1 ADO79627.1 ADO79628.1 ADO79629.1 ADO79630.1 ADO79631.1 KQ414666 KOC65098.1 HQ851372 AEV89750.1 JF770428 JF770430 AET83766.1 AET83768.1 JXUM01090826 JXUM01090827 JXUM01090828 KQ563868 KXJ73139.1 AXCM01002647
ABK60176.1 CAL69462.1 AY461705 AAR97568.1 AB290453 BAF73670.1 KT984953 AMY63172.1 KU549181 ANU04687.1 AGBW02008999 OWR51892.1 GAIX01003807 JAA88753.1 MG431817 AWX63640.1 KQ459595 KPI95936.1 AK401283 BAM17905.1 MH374163 AZK16057.1 JF811436 AEP43785.1 NWSH01001445 PCG71218.1 KQ460313 KPJ16050.1 MF797881 RSAL01000069 AWA45435.1 RVE49227.1 EU263634 ABX11259.1 ODYU01001173 SOQ37014.1 JQ009331 AFC98366.1 JTDY01000644 KOB76355.1 KZ149964 PZC76236.1 AB189024 BAD52256.1 AY460107 KA644700 AAR23787.1 AFP59329.1 KT270580 ALR99794.1 KT002138 KT002143 ALN12440.1 ALN12445.1 GFDF01000552 JAV13532.1 AJWK01029153 GFDG01001520 JAV17279.1 KT002146 ALN12448.1 JXJN01017347 DS235881 EEB20028.1 KF527568 AII26027.1 JXUM01073365 GAPW01005443 GAPW01005442 GAPW01005441 GAPW01005440 KQ562770 JAC08156.1 KXJ75156.1 GANO01001597 JAB58274.1 KP642166 ALM23457.1 MH006694 AVW80115.1 KP342269 KQ971339 AKH80544.1 EFA02790.1 DQ440061 CH477769 CH477190 ABF18094.1 EAT36424.1 EAT48702.1 MG601765 AXY54993.1 GEZM01037908 JAV81850.1 KC741174 AGM32998.1 CCAG010001380 EZ424081 ADD20357.1 GBYB01012041 JAG81808.1 JX263431 GAKP01008857 AGE89777.1 JAC50095.1 EZ617714 ADH94607.1 JRES01001549 KNC22106.1 KR072549 ALF39391.1 GDAI01001488 JAI16115.1 FX985484 BAX90092.1 GAMC01002843 JAC03713.1 M76975 NEVH01025158 PNF15606.1 GFDL01007346 JAV27699.1 MF776705 AVN67038.1 AY309973 AAQ81639.1 GBXI01012274 JAD02018.1 KK853022 KDR12362.1 KQ434829 KZC07670.1 GL437703 EFN70085.1 EF639098 GEMB01003510 ABR27983.1 JAR99707.1 AK417231 BAN20446.1 GBBI01000075 JAC18637.1 MG983778 AWV96613.1 GECL01003765 JAP02359.1 AY329355 AAP93581.1 GEBQ01028209 JAT11768.1 JR050197 KM244765 AEY61198.1 AIU41504.1 JN700517 KZ288318 AEZ57109.1 PBC28279.1 GDKW01000598 JAI55997.1 ACPB03006759 KQ976697 KYM77514.1 ADTU01020601 KK107238 QOIP01000007 EZA54646.1 RLU20399.1 DS232541 EDS43182.1 GEDC01023958 JAS13340.1 L13281 CH964101 CH916366 EDV97543.1 DQ096569 AAZ79896.1 X61687 CP012525 ALC44136.1 GBGD01003323 JAC85566.1 LNIX01000009 OXA49934.1 HQ230310 ADO20320.1 AB475105 BAH83704.1 GU300109 ADB54843.2 FJ554532 HQ326578 HQ326579 HQ326580 HQ326581 HQ326582 HQ326583 ACL80663.2 ADO79626.1 ADO79627.1 ADO79628.1 ADO79629.1 ADO79630.1 ADO79631.1 KQ414666 KOC65098.1 HQ851372 AEV89750.1 JF770428 JF770430 AET83766.1 AET83768.1 JXUM01090826 JXUM01090827 JXUM01090828 KQ563868 KXJ73139.1 AXCM01002647
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000053240
UP000283053
+ More
UP000037510 UP000095301 UP000092461 UP000095300 UP000092443 UP000092460 UP000009046 UP000192223 UP000078200 UP000069940 UP000249989 UP000092445 UP000007266 UP000008820 UP000092444 UP000037069 UP000235965 UP000091820 UP000027135 UP000076502 UP000000311 UP000005203 UP000242457 UP000015103 UP000078540 UP000005205 UP000053097 UP000279307 UP000002320 UP000007798 UP000001070 UP000079169 UP000092553 UP000198287 UP000053825 UP000075883
UP000037510 UP000095301 UP000092461 UP000095300 UP000092443 UP000092460 UP000009046 UP000192223 UP000078200 UP000069940 UP000249989 UP000092445 UP000007266 UP000008820 UP000092444 UP000037069 UP000235965 UP000091820 UP000027135 UP000076502 UP000000311 UP000005203 UP000242457 UP000015103 UP000078540 UP000005205 UP000053097 UP000279307 UP000002320 UP000007798 UP000001070 UP000079169 UP000092553 UP000198287 UP000053825 UP000075883
Pfam
PF00080 Sod_Cu
Interpro
SUPFAM
SSF49329
SSF49329
Gene 3D
ProteinModelPortal
Q5TLU4
A0JKF3
P82205
A7BI63
A0A170QHN4
A0A1B1RSA9
+ More
A0A212FDT2 S4PG22 A0A2Z4N444 A0A194PR57 I4DJ15 A0A3S5HJH2 G4XH81 A0A2A4JHM4 A0A194RFH4 A0A2S0RQR7 C3RSN1 A0A2H1V9V5 H9BEW2 A0A0L7LLQ0 A0A2W1BUB5 Q60FS5 A0A1I8N7L1 Q6SCL6 A0A0U2SX80 A0A0S2A460 A0A1L8E4S1 A0A1B0CUF9 A0A1L8EFB3 A0A0S2A496 A0A1I8Q6I1 A0A1A9XY61 A0A1B0BND7 E0W322 A0A1W4W7H5 A0A1A9UFW7 A0A076G467 A0A023EH27 U5EW44 A0A109WQA1 A0A1A9ZKK9 A0A2R4LGJ8 D2A2T2 Q1HRN3 A0A385HVY4 A0A1Y1MBB2 R4V538 D3TRM9 A0A0C9RW85 M1JNK3 F4MI33 A0A0L0BQ22 A0A0N7EJN0 A0A0K8TQF3 A0A3G9D4Q3 W8BWU5 P28755 A0A2J7PGY8 A0A1A9W501 A0A1Q3FJF2 A0A2P1H680 Q6W5R8 A0A0A1WUB5 A0A067R0S2 A0A154P938 E2A8Y6 A6YPT7 R4WNL0 A0A023FAY3 A0A2Z4G643 A0A0V0G2Z1 Q7YXM6 A0A088A933 A0A1B6KJY5 V9IJ95 H6UWL0 A0A0P4VTN9 T1HRT6 A0A195AZY4 A0A158NMI6 A0A026WG66 B0X9L3 A0A1B6CIU5 P41973 B4IY00 Q103C4 Q07182 A0A1S3D1W5 A0A0M5J141 A0A069DPF3 A0A226DYR0 E3VSS4 C5NS97 D2Y3W7 B8YNX4 A0A0L7R2N7 G9C5D1 G9D320 A0A182GVF6 A0A2C9GUA8
A0A212FDT2 S4PG22 A0A2Z4N444 A0A194PR57 I4DJ15 A0A3S5HJH2 G4XH81 A0A2A4JHM4 A0A194RFH4 A0A2S0RQR7 C3RSN1 A0A2H1V9V5 H9BEW2 A0A0L7LLQ0 A0A2W1BUB5 Q60FS5 A0A1I8N7L1 Q6SCL6 A0A0U2SX80 A0A0S2A460 A0A1L8E4S1 A0A1B0CUF9 A0A1L8EFB3 A0A0S2A496 A0A1I8Q6I1 A0A1A9XY61 A0A1B0BND7 E0W322 A0A1W4W7H5 A0A1A9UFW7 A0A076G467 A0A023EH27 U5EW44 A0A109WQA1 A0A1A9ZKK9 A0A2R4LGJ8 D2A2T2 Q1HRN3 A0A385HVY4 A0A1Y1MBB2 R4V538 D3TRM9 A0A0C9RW85 M1JNK3 F4MI33 A0A0L0BQ22 A0A0N7EJN0 A0A0K8TQF3 A0A3G9D4Q3 W8BWU5 P28755 A0A2J7PGY8 A0A1A9W501 A0A1Q3FJF2 A0A2P1H680 Q6W5R8 A0A0A1WUB5 A0A067R0S2 A0A154P938 E2A8Y6 A6YPT7 R4WNL0 A0A023FAY3 A0A2Z4G643 A0A0V0G2Z1 Q7YXM6 A0A088A933 A0A1B6KJY5 V9IJ95 H6UWL0 A0A0P4VTN9 T1HRT6 A0A195AZY4 A0A158NMI6 A0A026WG66 B0X9L3 A0A1B6CIU5 P41973 B4IY00 Q103C4 Q07182 A0A1S3D1W5 A0A0M5J141 A0A069DPF3 A0A226DYR0 E3VSS4 C5NS97 D2Y3W7 B8YNX4 A0A0L7R2N7 G9C5D1 G9D320 A0A182GVF6 A0A2C9GUA8
PDB
3L9E
E-value=2.94523e-74,
Score=702
Ontologies
PATHWAY
GO
PANTHER
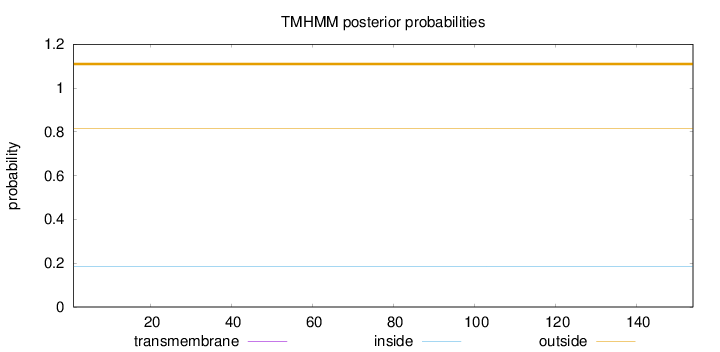
Topology
Subcellular location
Cytoplasm
Length:
154
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00053
Exp number, first 60 AAs:
0
Total prob of N-in:
0.18437
outside
1 - 154
Population Genetic Test Statistics
Pi
203.293713
Theta
167.220048
Tajima's D
0.712496
CLR
0.020045
CSRT
0.576171191440428
Interpretation
Uncertain
