Gene
KWMTBOMO13522
Pre Gene Modal
BGIBMGA001310
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_cytosol_aminopeptidase-like_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 2.144
Sequence
CDS
ATGAAGCTAGGCCAGGCGTTTGTACTGACCGACGTGGTCCCGGAGTACTCTGCTGTAGCGCTCGCTTCGTTAGGGCCAAAGGACCCTGGCTACAATCAGCTCGAAGCTTTGGATGAGACTAGGGAAAACCTTCGTTGGGCTGTTGGAGCTGGTGTGAGGGTCCTACAAAATCGAGGGTGTGGAGATATCTCCGTGCAGGCTGACGCGGAACACGACGCTGCAGCCGAGGCTGCCGAACTAGCCGCCTGGAGATTTGAAGAGTTCAGATCGGTAGAGGAGCGTCAGCCCGCGTGCAGGTTGTCGCTGCACGGGCAGGCGGGGGACGAGGGCCCCGGGGGCGCGTGGCACGCCGGCTCGGTGCTCGGTCGAGCGCAGAACTGGGCGAGGTTCTTGTCCGACATGCCCGCTAATAAAATGACTCCTATAGATCTTGGACAGGCAGCGTTAGACACGCTGTGTCCCCTGGGGGTGACCGTGGAAGCCAGAGAACGCGAGTGGATAGAGGCACAGAACATGCAGGCGTTCCTCACCGTAGCCCGAGGGTCCTGCGAGGAACCCATCTTCTTGGAATGTAATTACCGAGGAGACAAGGAATCTCGGCCTCCAGTCGTCATAGCCGCTAAAGGAGTTACTTTTGACAGCGGCGGCTTGTGTCTGAAGAAGCGGGAGAGCATGGTGGAGAACCGCGGCAGCATGGCCGGCGCCGCCGCCGTGCTGGGCGCGCTGCGGGCCCTCGCCGAGCTGCAGGTGCCTATCAATGTTTGCGCAGTCATACCGATATGCGAGAACATGGTGAGCGGACAGTGCATGAAGGTCGGAGACGTCGTCAGAGCTCTCAACGGACTCTCCATCCAGGTCCGAGGTCTTTGA
Protein
MKLGQAFVLTDVVPEYSAVALASLGPKDPGYNQLEALDETRENLRWAVGAGVRVLQNRGCGDISVQADAEHDAAAEAAELAAWRFEEFRSVEERQPACRLSLHGQAGDEGPGGAWHAGSVLGRAQNWARFLSDMPANKMTPIDLGQAALDTLCPLGVTVEAREREWIEAQNMQAFLTVARGSCEEPIFLECNYRGDKESRPPVVIAAKGVTFDSGGLCLKKRESMVENRGSMAGAAAVLGALRALAELQVPINVCAVIPICENMVSGQCMKVGDVVRALNGLSIQVRGL
Summary
Uniprot
H9IVM9
A0A2H1WCU5
A0A194PRI8
A0A212F8L7
A0A220K8P1
A0A182QM97
+ More
Q7PN16 W5J7E4 A0A1S4GWM4 A0A087ZI50 A0A182YGW9 A0A182JWX5 A0A182HVA6 A0A182WTB3 A0A182UU95 A0A182W9R8 A0A182NE59 R4G463 A0A182RBP4 A0A182F4I3 A0A182PMD8 A0A2P8YZJ3 A0A171AIP3 A0A3S2NJL2 A0A023ETB7 A0A182GA36 A0A023EUW2 Q173Y1 A0A224XEU8 B0W8W4 A0A1Q3FTI4 A0A161MNQ0 A0A023ESD8 A0A224XF43 A0A0A9VNX2 A0A224XM73 B4LKL0 A0A1I8M562 A0A0A9XHW0 T1I8S0 W8CAJ8 B4J580 A0A1B0AIZ5 A0A1A9WC21 A0A0A1XHN5 A0A171AIR8 A0A224XAZ3 A0A1I8P5K8 A0A2R7WK39 A0A023F7Y1 A0A034VYJ7 A0A0A9W468 A0A1B0G2B1 B3MFQ3 B4J7G8 A0A023F9A5 A0A171AIV7 A0A0M4EH04 A0A0L0CF64 B4KPY5 B4MSA4 A0A3B0J0N7 A0A336LS38 B4LKJ7 B4KSI3 A0A0A9YBR0 D6WAL5 A0A0M4EIQ0 R4G7V9 B5E1B9 B4GC96 A0A1S4MXI3 E0VYD5 A0A3B0J2T8 A0A0A9YMT8 B4J1L0 A0A336MSJ5 A0A1W4X7R3 B4GH68 Q28XX2 A0A0K8T3N8 B4LMF0 Q28XX6 B4PE18 B4MPH2 A0A1Y1KDF0 A0A3B0J0N1 B3NRH7 A0A1W4WWW8 B4N5V3 B3NPH6 A0A0J9RT45 B4HLV1 B4P658 Q7K2S9 B3NCL0 Q961W5 B4QI02 B4HSZ2 A0A2R7X258
Q7PN16 W5J7E4 A0A1S4GWM4 A0A087ZI50 A0A182YGW9 A0A182JWX5 A0A182HVA6 A0A182WTB3 A0A182UU95 A0A182W9R8 A0A182NE59 R4G463 A0A182RBP4 A0A182F4I3 A0A182PMD8 A0A2P8YZJ3 A0A171AIP3 A0A3S2NJL2 A0A023ETB7 A0A182GA36 A0A023EUW2 Q173Y1 A0A224XEU8 B0W8W4 A0A1Q3FTI4 A0A161MNQ0 A0A023ESD8 A0A224XF43 A0A0A9VNX2 A0A224XM73 B4LKL0 A0A1I8M562 A0A0A9XHW0 T1I8S0 W8CAJ8 B4J580 A0A1B0AIZ5 A0A1A9WC21 A0A0A1XHN5 A0A171AIR8 A0A224XAZ3 A0A1I8P5K8 A0A2R7WK39 A0A023F7Y1 A0A034VYJ7 A0A0A9W468 A0A1B0G2B1 B3MFQ3 B4J7G8 A0A023F9A5 A0A171AIV7 A0A0M4EH04 A0A0L0CF64 B4KPY5 B4MSA4 A0A3B0J0N7 A0A336LS38 B4LKJ7 B4KSI3 A0A0A9YBR0 D6WAL5 A0A0M4EIQ0 R4G7V9 B5E1B9 B4GC96 A0A1S4MXI3 E0VYD5 A0A3B0J2T8 A0A0A9YMT8 B4J1L0 A0A336MSJ5 A0A1W4X7R3 B4GH68 Q28XX2 A0A0K8T3N8 B4LMF0 Q28XX6 B4PE18 B4MPH2 A0A1Y1KDF0 A0A3B0J0N1 B3NRH7 A0A1W4WWW8 B4N5V3 B3NPH6 A0A0J9RT45 B4HLV1 B4P658 Q7K2S9 B3NCL0 Q961W5 B4QI02 B4HSZ2 A0A2R7X258
Pubmed
19121390
26354079
22118469
28630352
12364791
20920257
+ More
25244985 29403074 24945155 26483478 17510324 25401762 17994087 25315136 24495485 25830018 25474469 25348373 26108605 18362917 19820115 15632085 20566863 17550304 28004739 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
25244985 29403074 24945155 26483478 17510324 25401762 17994087 25315136 24495485 25830018 25474469 25348373 26108605 18362917 19820115 15632085 20566863 17550304 28004739 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01039650
BABH01039651
BABH01039652
BABH01039653
BABH01039654
BABH01039655
+ More
BABH01039656 BABH01039657 BABH01039658 ODYU01007736 SOQ50776.1 KQ459595 KPI95927.1 AGBW02009702 OWR50084.1 MF319730 ASJ26444.1 AXCN02000870 AAAB01008964 EAA12913.5 ADMH02002093 ETN59318.1 APCN01002503 ACPB03007327 GAHY01001615 JAA75895.1 PYGN01000268 PSN49667.1 GEMB01001016 JAS02130.1 RSAL01000069 RVE49219.1 GAPW01001023 JAC12575.1 JXUM01050664 KQ561657 KXJ77896.1 GAPW01000821 JAC12777.1 CH477415 EAT41391.1 GFTR01006892 JAW09534.1 DS231860 EDS39343.1 GFDL01004170 JAV30875.1 GEMB01001008 JAS02138.1 GAPW01001436 JAC12162.1 GFTR01006782 JAW09644.1 GBHO01045397 GBRD01009550 JAF98206.1 JAG56274.1 GFTR01006886 JAW09540.1 CH940648 EDW60731.1 GBHO01023287 GBRD01002795 JAG20317.1 JAG63026.1 ACPB03024616 GAMC01005676 JAC00880.1 CH916367 EDW00706.1 GBXI01003796 JAD10496.1 GEMB01001013 JAS02133.1 GFTR01006911 JAW09515.1 KK854966 PTY20004.1 GBBI01001350 JAC17362.1 GAKP01011448 JAC47504.1 GBHO01043969 GBRD01003915 JAF99634.1 JAG61906.1 CCAG010007175 CH902619 EDV37743.1 EDW01092.1 GBBI01001133 JAC17579.1 GEMB01001011 JAS02135.1 CP012525 ALC43790.1 JRES01000575 KNC30144.1 CH933808 EDW10262.1 CH963850 EDW74993.1 OUUW01000001 SPP74257.1 UFQT01000071 SSX19389.1 EDW60718.1 EDW09488.1 GBHO01015071 GBRD01010074 JAG28533.1 JAG55750.1 KQ971312 EEZ98647.1 CP012524 ALC41017.1 ACPB03019965 GAHY01002042 JAA75468.1 CM000071 EDY68724.1 CH479181 EDW31414.1 AAZO01006245 DS235845 EEB18391.1 SPP73453.1 GBHO01011191 JAG32413.1 CH916366 EDV96930.1 UFQS01002277 UFQT01002277 SSX13644.1 SSX33070.1 CH479183 EDW35838.1 EAL26194.2 GBRD01005669 JAG60152.1 EDW62045.1 EAL26190.2 CM000159 EDW94014.1 KRK01603.1 CH963849 EDW74011.1 GEZM01087697 JAV58528.1 SPP74247.1 CH954179 EDV56129.1 CH964154 EDW79742.1 EDV55743.1 CM002912 KMY98842.1 CH480815 EDW40983.1 CM000158 EDW91908.1 AE013599 AY060684 BT089026 AAF57988.1 AAL28232.1 ABV53819.1 ACU00259.1 CH954178 EDV51307.1 AY047518 AE014296 AAK77250.1 AAN11916.1 AAN11917.1 AGB94373.1 CM000362 CM002911 EDX07373.1 KMY94283.1 CH480816 EDW48156.1 KK856138 PTY24985.1
BABH01039656 BABH01039657 BABH01039658 ODYU01007736 SOQ50776.1 KQ459595 KPI95927.1 AGBW02009702 OWR50084.1 MF319730 ASJ26444.1 AXCN02000870 AAAB01008964 EAA12913.5 ADMH02002093 ETN59318.1 APCN01002503 ACPB03007327 GAHY01001615 JAA75895.1 PYGN01000268 PSN49667.1 GEMB01001016 JAS02130.1 RSAL01000069 RVE49219.1 GAPW01001023 JAC12575.1 JXUM01050664 KQ561657 KXJ77896.1 GAPW01000821 JAC12777.1 CH477415 EAT41391.1 GFTR01006892 JAW09534.1 DS231860 EDS39343.1 GFDL01004170 JAV30875.1 GEMB01001008 JAS02138.1 GAPW01001436 JAC12162.1 GFTR01006782 JAW09644.1 GBHO01045397 GBRD01009550 JAF98206.1 JAG56274.1 GFTR01006886 JAW09540.1 CH940648 EDW60731.1 GBHO01023287 GBRD01002795 JAG20317.1 JAG63026.1 ACPB03024616 GAMC01005676 JAC00880.1 CH916367 EDW00706.1 GBXI01003796 JAD10496.1 GEMB01001013 JAS02133.1 GFTR01006911 JAW09515.1 KK854966 PTY20004.1 GBBI01001350 JAC17362.1 GAKP01011448 JAC47504.1 GBHO01043969 GBRD01003915 JAF99634.1 JAG61906.1 CCAG010007175 CH902619 EDV37743.1 EDW01092.1 GBBI01001133 JAC17579.1 GEMB01001011 JAS02135.1 CP012525 ALC43790.1 JRES01000575 KNC30144.1 CH933808 EDW10262.1 CH963850 EDW74993.1 OUUW01000001 SPP74257.1 UFQT01000071 SSX19389.1 EDW60718.1 EDW09488.1 GBHO01015071 GBRD01010074 JAG28533.1 JAG55750.1 KQ971312 EEZ98647.1 CP012524 ALC41017.1 ACPB03019965 GAHY01002042 JAA75468.1 CM000071 EDY68724.1 CH479181 EDW31414.1 AAZO01006245 DS235845 EEB18391.1 SPP73453.1 GBHO01011191 JAG32413.1 CH916366 EDV96930.1 UFQS01002277 UFQT01002277 SSX13644.1 SSX33070.1 CH479183 EDW35838.1 EAL26194.2 GBRD01005669 JAG60152.1 EDW62045.1 EAL26190.2 CM000159 EDW94014.1 KRK01603.1 CH963849 EDW74011.1 GEZM01087697 JAV58528.1 SPP74247.1 CH954179 EDV56129.1 CH964154 EDW79742.1 EDV55743.1 CM002912 KMY98842.1 CH480815 EDW40983.1 CM000158 EDW91908.1 AE013599 AY060684 BT089026 AAF57988.1 AAL28232.1 ABV53819.1 ACU00259.1 CH954178 EDV51307.1 AY047518 AE014296 AAK77250.1 AAN11916.1 AAN11917.1 AGB94373.1 CM000362 CM002911 EDX07373.1 KMY94283.1 CH480816 EDW48156.1 KK856138 PTY24985.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000075886
UP000007062
UP000000673
+ More
UP000076408 UP000075881 UP000075840 UP000076407 UP000075903 UP000075920 UP000075884 UP000015103 UP000075900 UP000069272 UP000075885 UP000245037 UP000283053 UP000069940 UP000249989 UP000008820 UP000002320 UP000008792 UP000095301 UP000001070 UP000092445 UP000091820 UP000095300 UP000092444 UP000007801 UP000092553 UP000037069 UP000009192 UP000007798 UP000268350 UP000007266 UP000001819 UP000008744 UP000009046 UP000192223 UP000002282 UP000008711 UP000001292 UP000000803 UP000000304
UP000076408 UP000075881 UP000075840 UP000076407 UP000075903 UP000075920 UP000075884 UP000015103 UP000075900 UP000069272 UP000075885 UP000245037 UP000283053 UP000069940 UP000249989 UP000008820 UP000002320 UP000008792 UP000095301 UP000001070 UP000092445 UP000091820 UP000095300 UP000092444 UP000007801 UP000092553 UP000037069 UP000009192 UP000007798 UP000268350 UP000007266 UP000001819 UP000008744 UP000009046 UP000192223 UP000002282 UP000008711 UP000001292 UP000000803 UP000000304
Pfam
Interpro
IPR000819
Peptidase_M17_C
+ More
IPR037151 AlkB-like_sf
IPR036034 PDZ_sf
IPR008283 Peptidase_M17_N
IPR027450 AlkB-like
IPR001478 PDZ
IPR005123 Oxoglu/Fe-dep_dioxygenase
IPR041489 PDZ_6
IPR011356 Leucine_aapep/pepB
IPR019560 Mitochondrial_18_kDa_protein_
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR012934 Znf_AD
IPR035974 Rap/Ran-GAP_sf
IPR016024 ARM-type_fold
IPR000331 Rap_GAP_dom
IPR027107 Tuberin/Ral-act_asu
IPR037151 AlkB-like_sf
IPR036034 PDZ_sf
IPR008283 Peptidase_M17_N
IPR027450 AlkB-like
IPR001478 PDZ
IPR005123 Oxoglu/Fe-dep_dioxygenase
IPR041489 PDZ_6
IPR011356 Leucine_aapep/pepB
IPR019560 Mitochondrial_18_kDa_protein_
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR012934 Znf_AD
IPR035974 Rap/Ran-GAP_sf
IPR016024 ARM-type_fold
IPR000331 Rap_GAP_dom
IPR027107 Tuberin/Ral-act_asu
Gene 3D
ProteinModelPortal
H9IVM9
A0A2H1WCU5
A0A194PRI8
A0A212F8L7
A0A220K8P1
A0A182QM97
+ More
Q7PN16 W5J7E4 A0A1S4GWM4 A0A087ZI50 A0A182YGW9 A0A182JWX5 A0A182HVA6 A0A182WTB3 A0A182UU95 A0A182W9R8 A0A182NE59 R4G463 A0A182RBP4 A0A182F4I3 A0A182PMD8 A0A2P8YZJ3 A0A171AIP3 A0A3S2NJL2 A0A023ETB7 A0A182GA36 A0A023EUW2 Q173Y1 A0A224XEU8 B0W8W4 A0A1Q3FTI4 A0A161MNQ0 A0A023ESD8 A0A224XF43 A0A0A9VNX2 A0A224XM73 B4LKL0 A0A1I8M562 A0A0A9XHW0 T1I8S0 W8CAJ8 B4J580 A0A1B0AIZ5 A0A1A9WC21 A0A0A1XHN5 A0A171AIR8 A0A224XAZ3 A0A1I8P5K8 A0A2R7WK39 A0A023F7Y1 A0A034VYJ7 A0A0A9W468 A0A1B0G2B1 B3MFQ3 B4J7G8 A0A023F9A5 A0A171AIV7 A0A0M4EH04 A0A0L0CF64 B4KPY5 B4MSA4 A0A3B0J0N7 A0A336LS38 B4LKJ7 B4KSI3 A0A0A9YBR0 D6WAL5 A0A0M4EIQ0 R4G7V9 B5E1B9 B4GC96 A0A1S4MXI3 E0VYD5 A0A3B0J2T8 A0A0A9YMT8 B4J1L0 A0A336MSJ5 A0A1W4X7R3 B4GH68 Q28XX2 A0A0K8T3N8 B4LMF0 Q28XX6 B4PE18 B4MPH2 A0A1Y1KDF0 A0A3B0J0N1 B3NRH7 A0A1W4WWW8 B4N5V3 B3NPH6 A0A0J9RT45 B4HLV1 B4P658 Q7K2S9 B3NCL0 Q961W5 B4QI02 B4HSZ2 A0A2R7X258
Q7PN16 W5J7E4 A0A1S4GWM4 A0A087ZI50 A0A182YGW9 A0A182JWX5 A0A182HVA6 A0A182WTB3 A0A182UU95 A0A182W9R8 A0A182NE59 R4G463 A0A182RBP4 A0A182F4I3 A0A182PMD8 A0A2P8YZJ3 A0A171AIP3 A0A3S2NJL2 A0A023ETB7 A0A182GA36 A0A023EUW2 Q173Y1 A0A224XEU8 B0W8W4 A0A1Q3FTI4 A0A161MNQ0 A0A023ESD8 A0A224XF43 A0A0A9VNX2 A0A224XM73 B4LKL0 A0A1I8M562 A0A0A9XHW0 T1I8S0 W8CAJ8 B4J580 A0A1B0AIZ5 A0A1A9WC21 A0A0A1XHN5 A0A171AIR8 A0A224XAZ3 A0A1I8P5K8 A0A2R7WK39 A0A023F7Y1 A0A034VYJ7 A0A0A9W468 A0A1B0G2B1 B3MFQ3 B4J7G8 A0A023F9A5 A0A171AIV7 A0A0M4EH04 A0A0L0CF64 B4KPY5 B4MSA4 A0A3B0J0N7 A0A336LS38 B4LKJ7 B4KSI3 A0A0A9YBR0 D6WAL5 A0A0M4EIQ0 R4G7V9 B5E1B9 B4GC96 A0A1S4MXI3 E0VYD5 A0A3B0J2T8 A0A0A9YMT8 B4J1L0 A0A336MSJ5 A0A1W4X7R3 B4GH68 Q28XX2 A0A0K8T3N8 B4LMF0 Q28XX6 B4PE18 B4MPH2 A0A1Y1KDF0 A0A3B0J0N1 B3NRH7 A0A1W4WWW8 B4N5V3 B3NPH6 A0A0J9RT45 B4HLV1 B4P658 Q7K2S9 B3NCL0 Q961W5 B4QI02 B4HSZ2 A0A2R7X258
PDB
2EWB
E-value=1.18746e-40,
Score=417
Ontologies
KEGG
GO
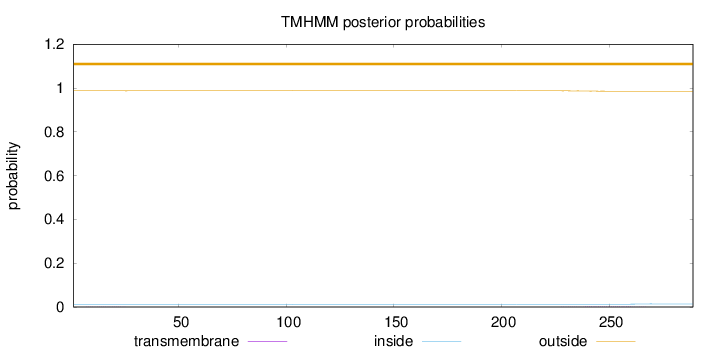
Topology
Length:
289
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05215
Exp number, first 60 AAs:
0.00622
Total prob of N-in:
0.01287
outside
1 - 289
Population Genetic Test Statistics
Pi
181.608912
Theta
150.355465
Tajima's D
0.723931
CLR
0.15854
CSRT
0.586920653967302
Interpretation
Uncertain
