Gene
KWMTBOMO13520 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001319
Annotation
Calmodulin?_partial_[Trichinella_pseudospiralis]
Location in the cell
Cytoplasmic Reliability : 3.413
Sequence
CDS
ATGGCGGATCAGCTGACGGAAGAACAGATCGCCGAGTTTAAGGAGGCATTCTCACTGTTCGACAAAGACGGCGATGGCACCATCACGACCAAAGAGCTGGGCACCGTGATGAGGTCGCTAGGACAGAACCCCACAGAAGCCGAACTTCAAGACATGATCAATGAAGTAGACGCGGACGGAAACGGCACGATAGACTTTCCCGAGTTCTTGACAATGATGGCGCGCAAGATGAAGGACACGGATAGCGAGGAAGAAATCCGCGAGGCCTTCCGCGTCTTCGACAAGGACGGCAACGGCTTCATCTCCGCGGCCGAGCTGCGCCACGTCATGACCAACCTCGGAGAGAAACTCACTGACGAGGAGGTCGACGAGATGATTCGCGAGGCCGATATCGACGGCGACGGCCAGGTCAATTACGAAGAGTTCGTCACCATGATGACGTCGAAGTGA
Protein
MADQLTEEQIAEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLTMMARKMKDTDSEEEIREAFRVFDKDGNGFISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEFVTMMTSK
Summary
Similarity
Belongs to the calmodulin family.
Uniprot
A0A2M4CHJ9
A0A0P6GTU5
A0A2M4CHG8
A0A2M4CI03
A0A1I9WLC8
A0A0V0XXP7
+ More
T1EAD2 A0A1L8E5R9 A0A171AUZ1 A0A2M3YY20 A0A1Q3FVD0 A0A0F7VHG0 A0A2L2Y0W1 A0A0F7VHE4 A0A0P5RUR5 C6SUZ2 A0A1B1X408 A0A1D1Z6U7 A0A1Y1LBB4 A0A224XZ95 D1FQ11 A0A1L8E5P3 T1D1N0 K4IPB7 A0A023EGV4 C1BZZ7 A0A224YTS4 W8BJI6 A0A2C9K5Z2 A0A221LCA4 R4SCH1 T1IQM0 A0A1W7RAV6 H8ZM86 A0A1W4X129 A0A0P4WF35 A0A3B0J5N8 Q6WSU5 A0A0N7Z9F5 A0A3G5APF9 T1E367 A0A1E1XJF4 A0A0B7A261 A0A131Z1F6 A0A3S0ZEV4 A0A1S3H7Q1 B4G9V3 K7IWY5 Q5XUA8 D6WB91 A0A0C9QZX5 A0A034WPE0 A0A0V0G4G1 A0A023F5W8 E7D1F3 Q76LB7 A0A0M3QV58 H6SWV2 A0A346QR72 E2J7D5 R4WCV1 A0A1J1IW19 A0A1D2N6J8 A0A088AD88 A0A075F404 A0A1S4H5D0 L7LXE1 Q1W2B3 R4S154 A0A023FWI6 I1V229 A0A0F7VJD4 I6LKW0 A0A3G5ANY4 A0A2I9LNT8 Q2F5T2 B4MY99 A0A293MVX1 A0A0L0C9L1 B4HP77 A0A1Z5LGF3 A0A131XC76 A0A0A1X0D7 S4PHH1 A0A191SEJ6 R9TI07 A0A2C9GQE4 A0A1B6DP62 A0A1E1X0D7 A0A1B6HBK9 A0A3L8DC46 E9H5Z2 K9S0T9 A0A069DN97 V4BWJ9 E3UJZ8 B4QC96 A0A1W4UHE8 A0A2J7QFD1 A0A0K8TWL1
T1EAD2 A0A1L8E5R9 A0A171AUZ1 A0A2M3YY20 A0A1Q3FVD0 A0A0F7VHG0 A0A2L2Y0W1 A0A0F7VHE4 A0A0P5RUR5 C6SUZ2 A0A1B1X408 A0A1D1Z6U7 A0A1Y1LBB4 A0A224XZ95 D1FQ11 A0A1L8E5P3 T1D1N0 K4IPB7 A0A023EGV4 C1BZZ7 A0A224YTS4 W8BJI6 A0A2C9K5Z2 A0A221LCA4 R4SCH1 T1IQM0 A0A1W7RAV6 H8ZM86 A0A1W4X129 A0A0P4WF35 A0A3B0J5N8 Q6WSU5 A0A0N7Z9F5 A0A3G5APF9 T1E367 A0A1E1XJF4 A0A0B7A261 A0A131Z1F6 A0A3S0ZEV4 A0A1S3H7Q1 B4G9V3 K7IWY5 Q5XUA8 D6WB91 A0A0C9QZX5 A0A034WPE0 A0A0V0G4G1 A0A023F5W8 E7D1F3 Q76LB7 A0A0M3QV58 H6SWV2 A0A346QR72 E2J7D5 R4WCV1 A0A1J1IW19 A0A1D2N6J8 A0A088AD88 A0A075F404 A0A1S4H5D0 L7LXE1 Q1W2B3 R4S154 A0A023FWI6 I1V229 A0A0F7VJD4 I6LKW0 A0A3G5ANY4 A0A2I9LNT8 Q2F5T2 B4MY99 A0A293MVX1 A0A0L0C9L1 B4HP77 A0A1Z5LGF3 A0A131XC76 A0A0A1X0D7 S4PHH1 A0A191SEJ6 R9TI07 A0A2C9GQE4 A0A1B6DP62 A0A1E1X0D7 A0A1B6HBK9 A0A3L8DC46 E9H5Z2 K9S0T9 A0A069DN97 V4BWJ9 E3UJZ8 B4QC96 A0A1W4UHE8 A0A2J7QFD1 A0A0K8TWL1
Pubmed
27538518
26561354
28004739
24945155
28797301
24495485
+ More
15562597 27717101 23671084 22348072 27129103 24330624 29209593 26830274 17994087 20075255 18362917 19820115 26131772 25348373 25474469 23691247 27289101 24997436 12364791 25576852 22507549 29248469 26108605 28528879 28049606 25830018 23622113 28503490 30249741 21292972 26334808 23254933 22044719 22936249
15562597 27717101 23671084 22348072 27129103 24330624 29209593 26830274 17994087 20075255 18362917 19820115 26131772 25348373 25474469 23691247 27289101 24997436 12364791 25576852 22507549 29248469 26108605 28528879 28049606 25830018 23622113 28503490 30249741 21292972 26334808 23254933 22044719 22936249
EMBL
GGFL01000527
MBW64705.1
GDIQ01028423
JAN66314.1
GGFL01000612
MBW64790.1
+ More
GGFL01000613 MBW64791.1 KU932312 APA33948.1 JYDU01000102 KRX92830.1 GAMD01000542 JAB01049.1 GFDF01000199 JAV13885.1 GEMB01000626 JAS02507.1 GGFM01000385 MBW21136.1 GFDL01003474 JAV31571.1 LN830645 CFW94154.1 IAAA01001390 IAAA01001391 IAAA01001392 LAA01836.1 LN830640 CFW94149.1 GDIQ01105206 JAL46520.1 BT088984 ACT88125.1 KU578024 ANW72275.1 GDJX01005340 JAT62596.1 GEZM01064027 JAV68856.1 GFTR01002514 JAW13912.1 EZ419911 ACZ28266.1 GFDF01000120 JAV13964.1 GAKT01000148 JAA92914.1 JX462608 AFU72271.1 GAPW01005554 JAC08044.1 BT080176 ACO14600.1 GFPF01006507 MAA17653.1 GAMC01013124 JAB93431.1 KX578900 ASM90733.1 KC810053 AGL94768.1 JH431312 GFAH01000116 JAV48273.1 JN314840 AFC93271.1 GDRN01035794 JAI67390.1 OUUW01000001 SPP75123.1 AY269783 AAQ01510.2 GDKW01000691 JAI55904.1 MH979776 AYV89231.1 GALA01000645 JAA94207.1 GFAA01004090 JAT99344.1 HACG01027336 CEK74201.1 GEDV01003303 JAP85254.1 RQTK01000695 RUS75976.1 CH479181 EDW31705.1 AAZX01000614 AAZX01004238 AY737546 AAU84939.1 KQ971312 EEZ98710.1 GBZX01002435 JAG90305.1 GAKP01002438 JAC56514.1 GECL01003178 JAP02946.1 GBBI01002137 JAC16575.1 HQ005901 ADV40197.1 AB081568 BAB89360.1 CP012524 ALC41839.1 HQ176458 ADX05545.1 MG910474 AXR98462.1 HP429353 ADN29853.1 AK417193 BAN20408.1 CVRI01000063 CRL04475.1 LJIJ01000184 ODN00851.1 KJ577632 AIE56158.1 AAAB01008823 GACK01008298 JAA56736.1 DQ445514 ABD98752.1 KC810055 AGL94770.1 GBBL01002222 JAC25098.1 JN688261 AFI25239.1 LN830702 CFW94211.2 GQ403698 ADF45338.1 MH990499 AYV89046.1 GFWZ01000061 MBW20051.1 DQ311341 ABD36285.1 CH963894 EDW77088.1 GFWV01020246 MAA44974.1 JRES01000735 KNC28895.1 CH480816 EDW47526.1 GFJQ02000483 JAW06487.1 GEFH01004012 JAP64569.1 GBXI01009971 JAD04321.1 GAIX01003277 JAA89283.1 KR733101 AND99539.1 KC989854 AGN29627.1 APCN01001422 GEDC01009849 JAS27449.1 GFAC01006481 JAT92707.1 GECU01035687 JAS72019.1 QOIP01000010 RLU17712.1 GL732595 EFX72807.1 JX855312 AFY63434.1 GBGD01003351 JAC85538.1 KB201931 ESO93369.1 HM445737 ADO64598.1 CM000362 CM002911 EDX06727.1 KMY93138.1 NEVH01015299 PNF27284.1 GDHF01033465 GDHF01028739 GDHF01020763 JAI18849.1 JAI23575.1 JAI31551.1
GGFL01000613 MBW64791.1 KU932312 APA33948.1 JYDU01000102 KRX92830.1 GAMD01000542 JAB01049.1 GFDF01000199 JAV13885.1 GEMB01000626 JAS02507.1 GGFM01000385 MBW21136.1 GFDL01003474 JAV31571.1 LN830645 CFW94154.1 IAAA01001390 IAAA01001391 IAAA01001392 LAA01836.1 LN830640 CFW94149.1 GDIQ01105206 JAL46520.1 BT088984 ACT88125.1 KU578024 ANW72275.1 GDJX01005340 JAT62596.1 GEZM01064027 JAV68856.1 GFTR01002514 JAW13912.1 EZ419911 ACZ28266.1 GFDF01000120 JAV13964.1 GAKT01000148 JAA92914.1 JX462608 AFU72271.1 GAPW01005554 JAC08044.1 BT080176 ACO14600.1 GFPF01006507 MAA17653.1 GAMC01013124 JAB93431.1 KX578900 ASM90733.1 KC810053 AGL94768.1 JH431312 GFAH01000116 JAV48273.1 JN314840 AFC93271.1 GDRN01035794 JAI67390.1 OUUW01000001 SPP75123.1 AY269783 AAQ01510.2 GDKW01000691 JAI55904.1 MH979776 AYV89231.1 GALA01000645 JAA94207.1 GFAA01004090 JAT99344.1 HACG01027336 CEK74201.1 GEDV01003303 JAP85254.1 RQTK01000695 RUS75976.1 CH479181 EDW31705.1 AAZX01000614 AAZX01004238 AY737546 AAU84939.1 KQ971312 EEZ98710.1 GBZX01002435 JAG90305.1 GAKP01002438 JAC56514.1 GECL01003178 JAP02946.1 GBBI01002137 JAC16575.1 HQ005901 ADV40197.1 AB081568 BAB89360.1 CP012524 ALC41839.1 HQ176458 ADX05545.1 MG910474 AXR98462.1 HP429353 ADN29853.1 AK417193 BAN20408.1 CVRI01000063 CRL04475.1 LJIJ01000184 ODN00851.1 KJ577632 AIE56158.1 AAAB01008823 GACK01008298 JAA56736.1 DQ445514 ABD98752.1 KC810055 AGL94770.1 GBBL01002222 JAC25098.1 JN688261 AFI25239.1 LN830702 CFW94211.2 GQ403698 ADF45338.1 MH990499 AYV89046.1 GFWZ01000061 MBW20051.1 DQ311341 ABD36285.1 CH963894 EDW77088.1 GFWV01020246 MAA44974.1 JRES01000735 KNC28895.1 CH480816 EDW47526.1 GFJQ02000483 JAW06487.1 GEFH01004012 JAP64569.1 GBXI01009971 JAD04321.1 GAIX01003277 JAA89283.1 KR733101 AND99539.1 KC989854 AGN29627.1 APCN01001422 GEDC01009849 JAS27449.1 GFAC01006481 JAT92707.1 GECU01035687 JAS72019.1 QOIP01000010 RLU17712.1 GL732595 EFX72807.1 JX855312 AFY63434.1 GBGD01003351 JAC85538.1 KB201931 ESO93369.1 HM445737 ADO64598.1 CM000362 CM002911 EDX06727.1 KMY93138.1 NEVH01015299 PNF27284.1 GDHF01033465 GDHF01028739 GDHF01020763 JAI18849.1 JAI23575.1 JAI31551.1
Proteomes
Pfam
PF13499 EF-hand_7
Interpro
SUPFAM
SSF47473
SSF47473
CDD
ProteinModelPortal
A0A2M4CHJ9
A0A0P6GTU5
A0A2M4CHG8
A0A2M4CI03
A0A1I9WLC8
A0A0V0XXP7
+ More
T1EAD2 A0A1L8E5R9 A0A171AUZ1 A0A2M3YY20 A0A1Q3FVD0 A0A0F7VHG0 A0A2L2Y0W1 A0A0F7VHE4 A0A0P5RUR5 C6SUZ2 A0A1B1X408 A0A1D1Z6U7 A0A1Y1LBB4 A0A224XZ95 D1FQ11 A0A1L8E5P3 T1D1N0 K4IPB7 A0A023EGV4 C1BZZ7 A0A224YTS4 W8BJI6 A0A2C9K5Z2 A0A221LCA4 R4SCH1 T1IQM0 A0A1W7RAV6 H8ZM86 A0A1W4X129 A0A0P4WF35 A0A3B0J5N8 Q6WSU5 A0A0N7Z9F5 A0A3G5APF9 T1E367 A0A1E1XJF4 A0A0B7A261 A0A131Z1F6 A0A3S0ZEV4 A0A1S3H7Q1 B4G9V3 K7IWY5 Q5XUA8 D6WB91 A0A0C9QZX5 A0A034WPE0 A0A0V0G4G1 A0A023F5W8 E7D1F3 Q76LB7 A0A0M3QV58 H6SWV2 A0A346QR72 E2J7D5 R4WCV1 A0A1J1IW19 A0A1D2N6J8 A0A088AD88 A0A075F404 A0A1S4H5D0 L7LXE1 Q1W2B3 R4S154 A0A023FWI6 I1V229 A0A0F7VJD4 I6LKW0 A0A3G5ANY4 A0A2I9LNT8 Q2F5T2 B4MY99 A0A293MVX1 A0A0L0C9L1 B4HP77 A0A1Z5LGF3 A0A131XC76 A0A0A1X0D7 S4PHH1 A0A191SEJ6 R9TI07 A0A2C9GQE4 A0A1B6DP62 A0A1E1X0D7 A0A1B6HBK9 A0A3L8DC46 E9H5Z2 K9S0T9 A0A069DN97 V4BWJ9 E3UJZ8 B4QC96 A0A1W4UHE8 A0A2J7QFD1 A0A0K8TWL1
T1EAD2 A0A1L8E5R9 A0A171AUZ1 A0A2M3YY20 A0A1Q3FVD0 A0A0F7VHG0 A0A2L2Y0W1 A0A0F7VHE4 A0A0P5RUR5 C6SUZ2 A0A1B1X408 A0A1D1Z6U7 A0A1Y1LBB4 A0A224XZ95 D1FQ11 A0A1L8E5P3 T1D1N0 K4IPB7 A0A023EGV4 C1BZZ7 A0A224YTS4 W8BJI6 A0A2C9K5Z2 A0A221LCA4 R4SCH1 T1IQM0 A0A1W7RAV6 H8ZM86 A0A1W4X129 A0A0P4WF35 A0A3B0J5N8 Q6WSU5 A0A0N7Z9F5 A0A3G5APF9 T1E367 A0A1E1XJF4 A0A0B7A261 A0A131Z1F6 A0A3S0ZEV4 A0A1S3H7Q1 B4G9V3 K7IWY5 Q5XUA8 D6WB91 A0A0C9QZX5 A0A034WPE0 A0A0V0G4G1 A0A023F5W8 E7D1F3 Q76LB7 A0A0M3QV58 H6SWV2 A0A346QR72 E2J7D5 R4WCV1 A0A1J1IW19 A0A1D2N6J8 A0A088AD88 A0A075F404 A0A1S4H5D0 L7LXE1 Q1W2B3 R4S154 A0A023FWI6 I1V229 A0A0F7VJD4 I6LKW0 A0A3G5ANY4 A0A2I9LNT8 Q2F5T2 B4MY99 A0A293MVX1 A0A0L0C9L1 B4HP77 A0A1Z5LGF3 A0A131XC76 A0A0A1X0D7 S4PHH1 A0A191SEJ6 R9TI07 A0A2C9GQE4 A0A1B6DP62 A0A1E1X0D7 A0A1B6HBK9 A0A3L8DC46 E9H5Z2 K9S0T9 A0A069DN97 V4BWJ9 E3UJZ8 B4QC96 A0A1W4UHE8 A0A2J7QFD1 A0A0K8TWL1
PDB
4IL1
E-value=4.41463e-76,
Score=718
Ontologies
PATHWAY
GO
GO:0005509
GO:0004683
GO:0046872
GO:0035071
GO:0072499
GO:0016028
GO:0007608
GO:0030048
GO:0006468
GO:0007605
GO:0030496
GO:0016060
GO:0016061
GO:0070855
GO:2001259
GO:0031476
GO:0046716
GO:0097431
GO:0007099
GO:0051383
GO:0071361
GO:0031475
GO:0005814
GO:0016056
GO:0005813
GO:0042052
GO:0031489
GO:0019722
GO:0016021
GO:0022857
GO:0051536
GO:0006777
GO:0005737
GO:0048869
GO:0003676
GO:0016491
GO:0015930
PANTHER
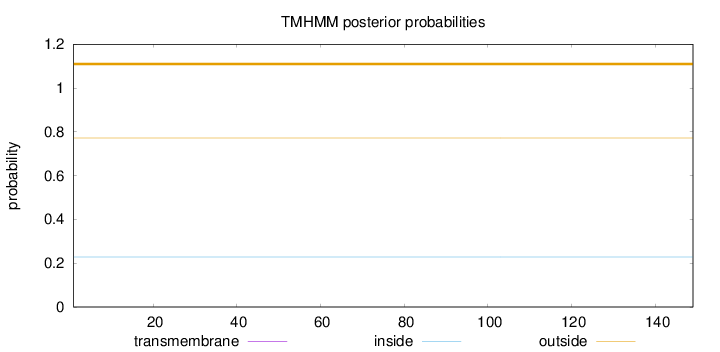
Topology
Length:
149
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.22849
outside
1 - 149
Population Genetic Test Statistics
Pi
20.022284
Theta
18.876476
Tajima's D
-0.936789
CLR
20.81194
CSRT
0.148442577871106
Interpretation
Uncertain
