Gene
KWMTBOMO13508
Pre Gene Modal
BGIBMGA009197
Annotation
PREDICTED:_organic_cation_transporter_protein-like_isoform_X1_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 0.885
Sequence
CDS
ATGTTCAGCATGACCGTGGTAGCTATGGAAGTATTCCCTACTAAGTCAAGAAACTCTCTCACTAATGTCGCCAACACCATCGGCAGGATTGGATCTATTTTAGCACCACAAGCACCATTACTGGAAAAATACATGAGTGGTCTCTCGAGCATCATTTTTGGAATAGTATCTTTGGGACCAGCTTTACTAGCATTCCTACTACCCGACACGAGTAAGGTTTCTCTACCAGACGACATCACAGAAGCAGAAAACTTAGACGTACGGCCGGAAGAATCTGACCCTACATGTAGTGTTCCTGCAAGTGGGGAATACAGGGAACGGTTTTGA
Protein
MFSMTVVAMEVFPTKSRNSLTNVANTIGRIGSILAPQAPLLEKYMSGLSSIIFGIVSLGPALLAFLLPDTSKVSLPDDITEAENLDVRPEESDPTCSVPASGEYRERF
Summary
Uniprot
H9JI49
A0A2H1W8C3
A0A2A4K7D4
A0A2W1BUD3
A0A194RER3
A0A194PXP4
+ More
H9JVZ7 A0A2W1BM98 H9JI51 A0A212F141 A0A2A4J3F4 A0A3S2L7U6 A0A2H1VQS6 A0A0L7LVB7 A0A0N0P9G0 A0A0N1I6I7 A0A0L7KUG4 A0A2A4JPL4 A0A212F2F2 A0A2H1W2A4 S4P1Y2 A0A2H1VDM1 A0A212EH35 A0A194RHT1 A0A194RH24 A0A194PCQ2 A0A212F072 A0A034UXN1 A0A2A4K6M9 A0A212EWR7 A0A212ERF4 A0A2A4K4I8 A0A194RI60 A0A0L7LSA4 A0A2H1V9B3 A0A2H1WTZ6 A0A2A4JU15 A0A2A4K3J3 A0A2A4K592 A0A3S2N7K6 A0A2R7VUS9 A0A2P8YSW5 A0A194RE15 A0A0N1PI45 A0A194QI00 A0A1B0DJC6 H9IWG9 A0A3S2L3J0 A0A3S2NA23 A0A194QC91 A0A212F2Q3 A0A2H1VA76 V9ICM7 A0A088AAF9 A0A2A3E3H4 A0A0L7RIG1 A0A2R7WSH8 A0A067RGL9 S4NXS3 W8BY90 A0A2J7RPZ6 H9IWG8 A0A194PCX0 A0A0J7KGE3 A0A154PCE9 A0A067RSY2 A0A2A4K5P1 A0A2J7RPZ7 B2DBH3 A0A182IUF7 A0A0L0C9F0 A0A1B6ML33 A0A194RIS2 A0A182SH91 A0A026W5E8 A0A182JXE2 A0A2J7RPY4 A0A195E462 A0A3L8D7U5 A0A2J7RPY2 A0A2H1V9G0 E2A7K4 H9IVN2 R4UKP6 E2BPX2 A0A0K8UFN6 A0A195FIT4 A0A2J7RPY0 A0A2J7RPY7 A0A0K8VEY5 A0A2A4K7E5 F4WUN7 A0A2J7RPY5 A0A0T6B887 B4L008 A0A067RDE7 A0A2A4K5R4 A0A2A4K7E3
H9JVZ7 A0A2W1BM98 H9JI51 A0A212F141 A0A2A4J3F4 A0A3S2L7U6 A0A2H1VQS6 A0A0L7LVB7 A0A0N0P9G0 A0A0N1I6I7 A0A0L7KUG4 A0A2A4JPL4 A0A212F2F2 A0A2H1W2A4 S4P1Y2 A0A2H1VDM1 A0A212EH35 A0A194RHT1 A0A194RH24 A0A194PCQ2 A0A212F072 A0A034UXN1 A0A2A4K6M9 A0A212EWR7 A0A212ERF4 A0A2A4K4I8 A0A194RI60 A0A0L7LSA4 A0A2H1V9B3 A0A2H1WTZ6 A0A2A4JU15 A0A2A4K3J3 A0A2A4K592 A0A3S2N7K6 A0A2R7VUS9 A0A2P8YSW5 A0A194RE15 A0A0N1PI45 A0A194QI00 A0A1B0DJC6 H9IWG9 A0A3S2L3J0 A0A3S2NA23 A0A194QC91 A0A212F2Q3 A0A2H1VA76 V9ICM7 A0A088AAF9 A0A2A3E3H4 A0A0L7RIG1 A0A2R7WSH8 A0A067RGL9 S4NXS3 W8BY90 A0A2J7RPZ6 H9IWG8 A0A194PCX0 A0A0J7KGE3 A0A154PCE9 A0A067RSY2 A0A2A4K5P1 A0A2J7RPZ7 B2DBH3 A0A182IUF7 A0A0L0C9F0 A0A1B6ML33 A0A194RIS2 A0A182SH91 A0A026W5E8 A0A182JXE2 A0A2J7RPY4 A0A195E462 A0A3L8D7U5 A0A2J7RPY2 A0A2H1V9G0 E2A7K4 H9IVN2 R4UKP6 E2BPX2 A0A0K8UFN6 A0A195FIT4 A0A2J7RPY0 A0A2J7RPY7 A0A0K8VEY5 A0A2A4K7E5 F4WUN7 A0A2J7RPY5 A0A0T6B887 B4L008 A0A067RDE7 A0A2A4K5R4 A0A2A4K7E3
Pubmed
EMBL
BABH01041119
BABH01041120
ODYU01006920
SOQ49196.1
NWSH01000100
PCG79560.1
+ More
KZ149964 PZC76256.1 KQ460313 KPJ16067.1 KQ459595 KPI95920.1 BABH01036481 KZ149965 PZC76202.1 BABH01041111 AGBW02010973 OWR47447.1 NWSH01003619 PCG66044.1 RSAL01000097 RVE47697.1 ODYU01003881 SOQ43193.1 JTDY01000022 KOB79380.1 KQ459449 KPJ00906.1 KQ461184 KPJ07510.1 JTDY01005496 KOB66937.1 NWSH01000834 PCG73941.1 AGBW02010753 OWR47904.1 ODYU01005874 SOQ47209.1 GAIX01012265 JAA80295.1 ODYU01001802 SOQ38502.1 AGBW02014973 OWR40799.1 KQ460205 KPJ17122.1 KPJ17123.1 KQ459606 KPI91071.1 AGBW02011137 OWR47156.1 GAKP01023245 JAC35711.1 PCG79564.1 AGBW02011950 OWR45874.1 AGBW02013021 OWR44080.1 NWSH01000168 PCG78824.1 KPJ17124.1 JTDY01000245 KOB78076.1 ODYU01001366 SOQ37440.1 ODYU01010990 SOQ56446.1 NWSH01000587 PCG75491.1 PCG78831.1 PCG78830.1 RSAL01002816 RVE40410.1 KK854099 PTY11252.1 PYGN01000379 PSN47342.1 KPJ16073.1 KQ459762 KPJ20130.1 KQ459193 KPJ03081.1 AJVK01064829 BABH01006656 RSAL01000186 RVE44802.1 RSAL01000360 RVE42197.1 KPJ03082.1 AGBW02010692 OWR48028.1 ODYU01001220 SOQ37134.1 JR038305 AEY58206.1 KZ288444 PBC25689.1 KQ414585 KOC70516.1 KK855422 PTY22512.1 KK852482 KDR22927.1 GAIX01010611 JAA81949.1 GAMC01012331 JAB94224.1 NEVH01001354 PNF42899.1 BABH01006651 BABH01006652 BABH01006653 KPI91072.1 LBMM01007950 KMQ89276.1 KQ434870 KZC09576.1 KDR22929.1 PCG79565.1 PNF42898.1 AB264653 BAG30732.1 JRES01000704 KNC29063.1 GEBQ01003309 JAT36668.1 KQ460325 KPJ15841.1 KK107403 EZA51300.1 PNF42897.1 KQ979657 KYN19975.1 QOIP01000011 RLU16525.1 PNF42895.1 SOQ37441.1 GL437348 EFN70576.1 BABH01039623 KC740880 AGM32704.1 GL449663 EFN82259.1 GDHF01026931 GDHF01013660 GDHF01009296 JAI25383.1 JAI38654.1 JAI43018.1 KQ981523 KYN40281.1 PNF42891.1 PNF42889.1 GDHF01014878 JAI37436.1 PCG79572.1 GL888375 EGI62034.1 PNF42894.1 LJIG01009268 KRT83419.1 CH933809 EDW19043.1 KK852530 KDR21906.1 PCG79571.1 PCG79570.1
KZ149964 PZC76256.1 KQ460313 KPJ16067.1 KQ459595 KPI95920.1 BABH01036481 KZ149965 PZC76202.1 BABH01041111 AGBW02010973 OWR47447.1 NWSH01003619 PCG66044.1 RSAL01000097 RVE47697.1 ODYU01003881 SOQ43193.1 JTDY01000022 KOB79380.1 KQ459449 KPJ00906.1 KQ461184 KPJ07510.1 JTDY01005496 KOB66937.1 NWSH01000834 PCG73941.1 AGBW02010753 OWR47904.1 ODYU01005874 SOQ47209.1 GAIX01012265 JAA80295.1 ODYU01001802 SOQ38502.1 AGBW02014973 OWR40799.1 KQ460205 KPJ17122.1 KPJ17123.1 KQ459606 KPI91071.1 AGBW02011137 OWR47156.1 GAKP01023245 JAC35711.1 PCG79564.1 AGBW02011950 OWR45874.1 AGBW02013021 OWR44080.1 NWSH01000168 PCG78824.1 KPJ17124.1 JTDY01000245 KOB78076.1 ODYU01001366 SOQ37440.1 ODYU01010990 SOQ56446.1 NWSH01000587 PCG75491.1 PCG78831.1 PCG78830.1 RSAL01002816 RVE40410.1 KK854099 PTY11252.1 PYGN01000379 PSN47342.1 KPJ16073.1 KQ459762 KPJ20130.1 KQ459193 KPJ03081.1 AJVK01064829 BABH01006656 RSAL01000186 RVE44802.1 RSAL01000360 RVE42197.1 KPJ03082.1 AGBW02010692 OWR48028.1 ODYU01001220 SOQ37134.1 JR038305 AEY58206.1 KZ288444 PBC25689.1 KQ414585 KOC70516.1 KK855422 PTY22512.1 KK852482 KDR22927.1 GAIX01010611 JAA81949.1 GAMC01012331 JAB94224.1 NEVH01001354 PNF42899.1 BABH01006651 BABH01006652 BABH01006653 KPI91072.1 LBMM01007950 KMQ89276.1 KQ434870 KZC09576.1 KDR22929.1 PCG79565.1 PNF42898.1 AB264653 BAG30732.1 JRES01000704 KNC29063.1 GEBQ01003309 JAT36668.1 KQ460325 KPJ15841.1 KK107403 EZA51300.1 PNF42897.1 KQ979657 KYN19975.1 QOIP01000011 RLU16525.1 PNF42895.1 SOQ37441.1 GL437348 EFN70576.1 BABH01039623 KC740880 AGM32704.1 GL449663 EFN82259.1 GDHF01026931 GDHF01013660 GDHF01009296 JAI25383.1 JAI38654.1 JAI43018.1 KQ981523 KYN40281.1 PNF42891.1 PNF42889.1 GDHF01014878 JAI37436.1 PCG79572.1 GL888375 EGI62034.1 PNF42894.1 LJIG01009268 KRT83419.1 CH933809 EDW19043.1 KK852530 KDR21906.1 PCG79571.1 PCG79570.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000283053
+ More
UP000037510 UP000245037 UP000092462 UP000005203 UP000242457 UP000053825 UP000027135 UP000235965 UP000036403 UP000076502 UP000075880 UP000037069 UP000075901 UP000053097 UP000075881 UP000078492 UP000279307 UP000000311 UP000008237 UP000078541 UP000007755 UP000009192
UP000037510 UP000245037 UP000092462 UP000005203 UP000242457 UP000053825 UP000027135 UP000235965 UP000036403 UP000076502 UP000075880 UP000037069 UP000075901 UP000053097 UP000075881 UP000078492 UP000279307 UP000000311 UP000008237 UP000078541 UP000007755 UP000009192
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JI49
A0A2H1W8C3
A0A2A4K7D4
A0A2W1BUD3
A0A194RER3
A0A194PXP4
+ More
H9JVZ7 A0A2W1BM98 H9JI51 A0A212F141 A0A2A4J3F4 A0A3S2L7U6 A0A2H1VQS6 A0A0L7LVB7 A0A0N0P9G0 A0A0N1I6I7 A0A0L7KUG4 A0A2A4JPL4 A0A212F2F2 A0A2H1W2A4 S4P1Y2 A0A2H1VDM1 A0A212EH35 A0A194RHT1 A0A194RH24 A0A194PCQ2 A0A212F072 A0A034UXN1 A0A2A4K6M9 A0A212EWR7 A0A212ERF4 A0A2A4K4I8 A0A194RI60 A0A0L7LSA4 A0A2H1V9B3 A0A2H1WTZ6 A0A2A4JU15 A0A2A4K3J3 A0A2A4K592 A0A3S2N7K6 A0A2R7VUS9 A0A2P8YSW5 A0A194RE15 A0A0N1PI45 A0A194QI00 A0A1B0DJC6 H9IWG9 A0A3S2L3J0 A0A3S2NA23 A0A194QC91 A0A212F2Q3 A0A2H1VA76 V9ICM7 A0A088AAF9 A0A2A3E3H4 A0A0L7RIG1 A0A2R7WSH8 A0A067RGL9 S4NXS3 W8BY90 A0A2J7RPZ6 H9IWG8 A0A194PCX0 A0A0J7KGE3 A0A154PCE9 A0A067RSY2 A0A2A4K5P1 A0A2J7RPZ7 B2DBH3 A0A182IUF7 A0A0L0C9F0 A0A1B6ML33 A0A194RIS2 A0A182SH91 A0A026W5E8 A0A182JXE2 A0A2J7RPY4 A0A195E462 A0A3L8D7U5 A0A2J7RPY2 A0A2H1V9G0 E2A7K4 H9IVN2 R4UKP6 E2BPX2 A0A0K8UFN6 A0A195FIT4 A0A2J7RPY0 A0A2J7RPY7 A0A0K8VEY5 A0A2A4K7E5 F4WUN7 A0A2J7RPY5 A0A0T6B887 B4L008 A0A067RDE7 A0A2A4K5R4 A0A2A4K7E3
H9JVZ7 A0A2W1BM98 H9JI51 A0A212F141 A0A2A4J3F4 A0A3S2L7U6 A0A2H1VQS6 A0A0L7LVB7 A0A0N0P9G0 A0A0N1I6I7 A0A0L7KUG4 A0A2A4JPL4 A0A212F2F2 A0A2H1W2A4 S4P1Y2 A0A2H1VDM1 A0A212EH35 A0A194RHT1 A0A194RH24 A0A194PCQ2 A0A212F072 A0A034UXN1 A0A2A4K6M9 A0A212EWR7 A0A212ERF4 A0A2A4K4I8 A0A194RI60 A0A0L7LSA4 A0A2H1V9B3 A0A2H1WTZ6 A0A2A4JU15 A0A2A4K3J3 A0A2A4K592 A0A3S2N7K6 A0A2R7VUS9 A0A2P8YSW5 A0A194RE15 A0A0N1PI45 A0A194QI00 A0A1B0DJC6 H9IWG9 A0A3S2L3J0 A0A3S2NA23 A0A194QC91 A0A212F2Q3 A0A2H1VA76 V9ICM7 A0A088AAF9 A0A2A3E3H4 A0A0L7RIG1 A0A2R7WSH8 A0A067RGL9 S4NXS3 W8BY90 A0A2J7RPZ6 H9IWG8 A0A194PCX0 A0A0J7KGE3 A0A154PCE9 A0A067RSY2 A0A2A4K5P1 A0A2J7RPZ7 B2DBH3 A0A182IUF7 A0A0L0C9F0 A0A1B6ML33 A0A194RIS2 A0A182SH91 A0A026W5E8 A0A182JXE2 A0A2J7RPY4 A0A195E462 A0A3L8D7U5 A0A2J7RPY2 A0A2H1V9G0 E2A7K4 H9IVN2 R4UKP6 E2BPX2 A0A0K8UFN6 A0A195FIT4 A0A2J7RPY0 A0A2J7RPY7 A0A0K8VEY5 A0A2A4K7E5 F4WUN7 A0A2J7RPY5 A0A0T6B887 B4L008 A0A067RDE7 A0A2A4K5R4 A0A2A4K7E3
Ontologies
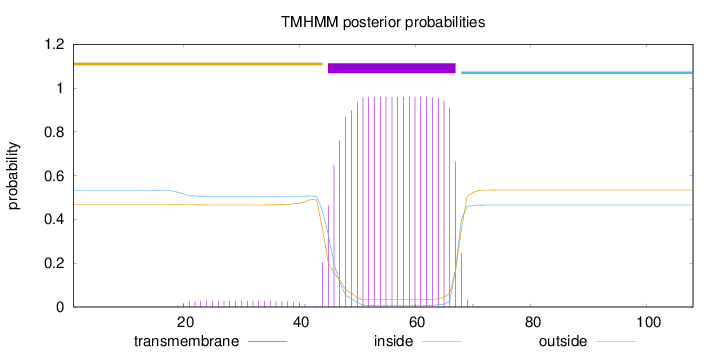
Topology
Length:
108
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.59951
Exp number, first 60 AAs:
14.95524
Total prob of N-in:
0.53142
POSSIBLE N-term signal
sequence
outside
1 - 44
TMhelix
45 - 67
inside
68 - 108
Population Genetic Test Statistics
Pi
213.624433
Theta
178.799344
Tajima's D
0.952433
CLR
0
CSRT
0.645817709114544
Interpretation
Uncertain
