Gene
KWMTBOMO13506
Pre Gene Modal
BGIBMGA009201
Annotation
PREDICTED:_organic_cation_transporter_protein-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.737
Sequence
CDS
ATGGCAGACAGAAAACCCAGCGTGGACCCCGCGAGGAAGGGCAGCGCATTTGACCAGCCAATAGATCTAGACCATGTGCTCATCAACGAGCTGGGACAGTTTGGATTGTTTCAGTTGAGGAACATCCTCTTGGTGGCCATACCTATTATCATGTCGGCTTTCATGAGCGAATTCGTCTTTTCTGCTGCAGCTATTCCTCACAGGTGTCGGATCCCAGAGTGCGGGGAAGTTGAGAAAAGTACGAATTACGACCGAGAGTGGTTGGCGAACGCCATCCCTGAGAGCGACTCTGGATTCGCGAGTTGTGAGCGGTTCGCTCCAGTCACCGGGATCAACGGGACATTGGACTACTGTCCGCCGAATATCTTCCAGACCAACACCATTGAGTGCGATGGATTCATTTATACCAGAGATAACTCTGTTGTTTTTGACTTCGACCTTGGGTGCCAGGAGTGGCTTCGTGCTCTTGCCGGCACTTTATCCAGCGTGGGGACATTGCTGGTCCTGCCCATCACTGGTTATGTTTCCGACCGCTTCGGGAGACGTATGGCCCTAGTGATCAGCGTCTTCAACTTGGCACTATTTGGCTTAATCAAAGCCTTCTCCGTAAACTACGTTATGTACTTGGCTTTCCAGGTAATACAGACGACTCTGGGGGCCGGCACCTTCAGCTCCGCCTACATATTCGCCACTGAATTCGTGGGACCGAAGTACCGAGTCATTACCAGTGCGACCTGCTCATCGATGTTCGCGGTCGGTCAAGTGGTCCTCGGGGGGATCGCGTGGCTGGTCCAGCCCTGGCGGTACATGATCATGACGCTCCACATCCCTTGTTTCCTTATCGTCTCATACTACTGGCTACTATCGGAGAGCGTAAGATGGCTGCTCTCAAAGAGAAAATTCGATGAAGCCCGACGGGTCTTGGAGAAAGCTGCCCATGTTAACAAGAGAGAGATTAGTCAGAAGTCTATGAACGCATTGATGAGGCCTCCAGCGGCGGTCGCCACGGCGGAACCCGTAGAAGCAGACCCTGGAGTTCTCAGAACCATAATCAAATCTCCTATCCTACTCCGTCGTGTGTGCACCACGCCGATCTGGTGGATCACCACGACCTTCGTGTACTACGGCCTCTCCATCAACTCCGCCAGCTTATCAGACACCATTTACTTGAACTACATCCTGACCTGTGCTATCGAAATCCCTGGATTTTACACTGCGGTCTTGATTCTGGACAGGGTCGGAAGAAAGCCGACTTTATCTGGTGGATTCTTCTTCAGTGCTCTTTGCAATATAGCCTTTGCTTTCATACCTAACGACATGACAGTTCTTCGCTTGATAATATTCCTTCTGGGCAAGTTCGGTATCTCCATGGTGTTCACATCCCTGTACCTGTTCACGTCCGAGCTGTACCCCACGCAGTACAGACACAGCCTGCTCGCATTCTCGTCCATGATCGGTCGCATCGGATCCATCACTGCTCCACTCACTCCTGCTTTAATGGAGTATTGGCATGGCATACCTACAGTGATGTTCGGCGCTATGGGCTTCCTCTCCGGAGTGCTGATCCTCACTCAGCCCGAAACCCTGGGCACCAAGATGCCGAACACACTCGCAGAGGCAGAGGCGCTTGGCACTCCTCAATCCAAGATACAAAGCAACTAA
Protein
MADRKPSVDPARKGSAFDQPIDLDHVLINELGQFGLFQLRNILLVAIPIIMSAFMSEFVFSAAAIPHRCRIPECGEVEKSTNYDREWLANAIPESDSGFASCERFAPVTGINGTLDYCPPNIFQTNTIECDGFIYTRDNSVVFDFDLGCQEWLRALAGTLSSVGTLLVLPITGYVSDRFGRRMALVISVFNLALFGLIKAFSVNYVMYLAFQVIQTTLGAGTFSSAYIFATEFVGPKYRVITSATCSSMFAVGQVVLGGIAWLVQPWRYMIMTLHIPCFLIVSYYWLLSESVRWLLSKRKFDEARRVLEKAAHVNKREISQKSMNALMRPPAAVATAEPVEADPGVLRTIIKSPILLRRVCTTPIWWITTTFVYYGLSINSASLSDTIYLNYILTCAIEIPGFYTAVLILDRVGRKPTLSGGFFFSALCNIAFAFIPNDMTVLRLIIFLLGKFGISMVFTSLYLFTSELYPTQYRHSLLAFSSMIGRIGSITAPLTPALMEYWHGIPTVMFGAMGFLSGVLILTQPETLGTKMPNTLAEAEALGTPQSKIQSN
Summary
Uniprot
H9JI53
H9JI54
A0A0L7KV39
A0A194PRI3
A0A2H1V9D8
A0A3S2NQS4
+ More
A0A2A4K5Q3 A0A2W1BRL6 A0A3S2TCP3 A0A212F123 A0A194PR44 A0A212F124 A0A194RF38 A0A0L7KZ57 A0A194RJF2 H9JI52 A0A2A4J6J0 A0A0L7KWH4 A0A194RGD2 A0A212F137 A0A194PJI6 A0A194RF43 A0A2A4K5Q2 A0A3S2L9U7 A0A212EJ24 A0A212EMB5 A0A2W1BMH3 A0A0L7KTQ3 A0A194RE10 A0A194PT27 A0A2H1VQB3 A0A194PSH3 A0A2A4K663 A0A2H1VCJ6 A0A2H1VQC6 A0A2W1BSR9 A0A2A4JIA3 A0A2H1VAT9 A0A2W1BHB5 A0A2A4JH68 A0A2W1BLR2 A0A3S2LQC2 A0A2H1VSF4 A0A0L7K3V5 A0A2A4K6L7 A0A0L7KTW1 A0A2W1BMV0 A0A2H1V9B3 H9JI48 A0A3S2NA23 A0A0L7KVH9 A0A0L7K4R7 A0A194QDP3 A0A212FPC5 A0A3S2NDN7 A0A182SYI8 B0XDF1 A0A2J7RPZ7 Q7Q0C3 Q16WK7 A0A2A4K6M9 B0XDF0 A0A2J7RPY7 A0A182LZQ5 A0A2J7RPY0 A0A067RSY2 A0A3S2M6B8 A0A212EJ20 A0A2H1VA76 A0A182Y1J3 A0A3S2NIQ2 A0A182QUL1 A0A182GYP2 A0A3S2L3J0 A0A1L8DEQ3 A0A1L8DEI6 A0A194QI00 A0A2A4J5Q3 A0A2H1VFS1 A0A182KSD5 A0A194PT22 A0A194QC91 A0A2W1BRJ1 A0A2A4JC88 A0A0N1PI45 A0A0L7LKI8 A0A194RIS2 A0A212F2Q3 A0A182MYV4 A0A2A4K5R4 A0A2A4K5P1 A0A182RFF8 A0A195E462 B0XDE9 A0A0K8VUC5 A0A1Y1KD84 A0A0K8UFN6
A0A2A4K5Q3 A0A2W1BRL6 A0A3S2TCP3 A0A212F123 A0A194PR44 A0A212F124 A0A194RF38 A0A0L7KZ57 A0A194RJF2 H9JI52 A0A2A4J6J0 A0A0L7KWH4 A0A194RGD2 A0A212F137 A0A194PJI6 A0A194RF43 A0A2A4K5Q2 A0A3S2L9U7 A0A212EJ24 A0A212EMB5 A0A2W1BMH3 A0A0L7KTQ3 A0A194RE10 A0A194PT27 A0A2H1VQB3 A0A194PSH3 A0A2A4K663 A0A2H1VCJ6 A0A2H1VQC6 A0A2W1BSR9 A0A2A4JIA3 A0A2H1VAT9 A0A2W1BHB5 A0A2A4JH68 A0A2W1BLR2 A0A3S2LQC2 A0A2H1VSF4 A0A0L7K3V5 A0A2A4K6L7 A0A0L7KTW1 A0A2W1BMV0 A0A2H1V9B3 H9JI48 A0A3S2NA23 A0A0L7KVH9 A0A0L7K4R7 A0A194QDP3 A0A212FPC5 A0A3S2NDN7 A0A182SYI8 B0XDF1 A0A2J7RPZ7 Q7Q0C3 Q16WK7 A0A2A4K6M9 B0XDF0 A0A2J7RPY7 A0A182LZQ5 A0A2J7RPY0 A0A067RSY2 A0A3S2M6B8 A0A212EJ20 A0A2H1VA76 A0A182Y1J3 A0A3S2NIQ2 A0A182QUL1 A0A182GYP2 A0A3S2L3J0 A0A1L8DEQ3 A0A1L8DEI6 A0A194QI00 A0A2A4J5Q3 A0A2H1VFS1 A0A182KSD5 A0A194PT22 A0A194QC91 A0A2W1BRJ1 A0A2A4JC88 A0A0N1PI45 A0A0L7LKI8 A0A194RIS2 A0A212F2Q3 A0A182MYV4 A0A2A4K5R4 A0A2A4K5P1 A0A182RFF8 A0A195E462 B0XDE9 A0A0K8VUC5 A0A1Y1KD84 A0A0K8UFN6
Pubmed
EMBL
BABH01041117
BABH01041118
BABH01041119
JTDY01005496
KOB66936.1
KQ459595
+ More
KPI95922.1 ODYU01001366 SOQ37439.1 RSAL01000360 RVE42198.1 NWSH01000100 PCG79561.1 KZ149964 PZC76255.1 RVE42200.1 AGBW02010973 OWR47448.1 KPI95921.1 OWR47449.1 KQ460313 KPJ16064.1 JTDY01004270 KOB68356.1 KPJ16066.1 BABH01041114 NWSH01002815 PCG67489.1 JTDY01005018 KOB67470.1 KQ460205 KPJ16878.1 OWR47450.1 KQ459606 KPI91250.1 KPJ16069.1 PCG79557.1 RVE42203.1 AGBW02014542 OWR41479.1 AGBW02013857 OWR42632.1 PZC76259.1 JTDY01005909 KOB66490.1 KPJ16068.1 KPI95919.1 ODYU01003788 SOQ42987.1 KPI95918.1 PCG79559.1 ODYU01001566 SOQ37954.1 SOQ42986.1 PZC76257.1 NWSH01001466 PCG71160.1 SOQ37955.1 KZ150061 PZC74228.1 PCG71159.1 PZC74227.1 RVE42202.1 ODYU01004183 SOQ43775.1 JTDY01011376 KOB56709.1 PCG79558.1 KOB66489.1 PZC76258.1 SOQ37440.1 BABH01041122 RVE42197.1 JTDY01005309 KOB67130.1 JTDY01010709 KOB58080.1 KQ459193 KPJ03085.1 AGBW02003000 OWR55596.1 RSAL01000186 RVE44804.1 DS232756 EDS45429.1 NEVH01001354 PNF42898.1 AAAB01008986 EAA00192.3 CH477563 EAT38951.1 PCG79564.1 EDS45428.1 PNF42889.1 AXCM01003135 PNF42891.1 KK852482 KDR22929.1 RSAL01000027 RVE51971.1 OWR41475.1 ODYU01001220 SOQ37134.1 RSAL01000974 RVE41008.1 AXCN02001021 JXUM01021216 KQ560594 KXJ81736.1 RVE44802.1 GFDF01009227 JAV04857.1 GFDF01009228 JAV04856.1 KPJ03081.1 NWSH01003151 PCG66854.1 ODYU01002328 SOQ39679.1 KPI95914.1 KPJ03082.1 PZC76264.1 NWSH01002161 PCG69013.1 KQ459762 KPJ20130.1 JTDY01000842 KOB75696.1 KQ460325 KPJ15841.1 AGBW02010692 OWR48028.1 PCG79571.1 PCG79565.1 KQ979657 KYN19975.1 EDS45427.1 GDHF01009833 JAI42481.1 GEZM01087818 JAV58458.1 GDHF01026931 GDHF01013660 GDHF01009296 JAI25383.1 JAI38654.1 JAI43018.1
KPI95922.1 ODYU01001366 SOQ37439.1 RSAL01000360 RVE42198.1 NWSH01000100 PCG79561.1 KZ149964 PZC76255.1 RVE42200.1 AGBW02010973 OWR47448.1 KPI95921.1 OWR47449.1 KQ460313 KPJ16064.1 JTDY01004270 KOB68356.1 KPJ16066.1 BABH01041114 NWSH01002815 PCG67489.1 JTDY01005018 KOB67470.1 KQ460205 KPJ16878.1 OWR47450.1 KQ459606 KPI91250.1 KPJ16069.1 PCG79557.1 RVE42203.1 AGBW02014542 OWR41479.1 AGBW02013857 OWR42632.1 PZC76259.1 JTDY01005909 KOB66490.1 KPJ16068.1 KPI95919.1 ODYU01003788 SOQ42987.1 KPI95918.1 PCG79559.1 ODYU01001566 SOQ37954.1 SOQ42986.1 PZC76257.1 NWSH01001466 PCG71160.1 SOQ37955.1 KZ150061 PZC74228.1 PCG71159.1 PZC74227.1 RVE42202.1 ODYU01004183 SOQ43775.1 JTDY01011376 KOB56709.1 PCG79558.1 KOB66489.1 PZC76258.1 SOQ37440.1 BABH01041122 RVE42197.1 JTDY01005309 KOB67130.1 JTDY01010709 KOB58080.1 KQ459193 KPJ03085.1 AGBW02003000 OWR55596.1 RSAL01000186 RVE44804.1 DS232756 EDS45429.1 NEVH01001354 PNF42898.1 AAAB01008986 EAA00192.3 CH477563 EAT38951.1 PCG79564.1 EDS45428.1 PNF42889.1 AXCM01003135 PNF42891.1 KK852482 KDR22929.1 RSAL01000027 RVE51971.1 OWR41475.1 ODYU01001220 SOQ37134.1 RSAL01000974 RVE41008.1 AXCN02001021 JXUM01021216 KQ560594 KXJ81736.1 RVE44802.1 GFDF01009227 JAV04857.1 GFDF01009228 JAV04856.1 KPJ03081.1 NWSH01003151 PCG66854.1 ODYU01002328 SOQ39679.1 KPI95914.1 KPJ03082.1 PZC76264.1 NWSH01002161 PCG69013.1 KQ459762 KPJ20130.1 JTDY01000842 KOB75696.1 KQ460325 KPJ15841.1 AGBW02010692 OWR48028.1 PCG79571.1 PCG79565.1 KQ979657 KYN19975.1 EDS45427.1 GDHF01009833 JAI42481.1 GEZM01087818 JAV58458.1 GDHF01026931 GDHF01013660 GDHF01009296 JAI25383.1 JAI38654.1 JAI43018.1
Proteomes
Interpro
SUPFAM
SSF103473
SSF103473
Gene 3D
CDD
ProteinModelPortal
H9JI53
H9JI54
A0A0L7KV39
A0A194PRI3
A0A2H1V9D8
A0A3S2NQS4
+ More
A0A2A4K5Q3 A0A2W1BRL6 A0A3S2TCP3 A0A212F123 A0A194PR44 A0A212F124 A0A194RF38 A0A0L7KZ57 A0A194RJF2 H9JI52 A0A2A4J6J0 A0A0L7KWH4 A0A194RGD2 A0A212F137 A0A194PJI6 A0A194RF43 A0A2A4K5Q2 A0A3S2L9U7 A0A212EJ24 A0A212EMB5 A0A2W1BMH3 A0A0L7KTQ3 A0A194RE10 A0A194PT27 A0A2H1VQB3 A0A194PSH3 A0A2A4K663 A0A2H1VCJ6 A0A2H1VQC6 A0A2W1BSR9 A0A2A4JIA3 A0A2H1VAT9 A0A2W1BHB5 A0A2A4JH68 A0A2W1BLR2 A0A3S2LQC2 A0A2H1VSF4 A0A0L7K3V5 A0A2A4K6L7 A0A0L7KTW1 A0A2W1BMV0 A0A2H1V9B3 H9JI48 A0A3S2NA23 A0A0L7KVH9 A0A0L7K4R7 A0A194QDP3 A0A212FPC5 A0A3S2NDN7 A0A182SYI8 B0XDF1 A0A2J7RPZ7 Q7Q0C3 Q16WK7 A0A2A4K6M9 B0XDF0 A0A2J7RPY7 A0A182LZQ5 A0A2J7RPY0 A0A067RSY2 A0A3S2M6B8 A0A212EJ20 A0A2H1VA76 A0A182Y1J3 A0A3S2NIQ2 A0A182QUL1 A0A182GYP2 A0A3S2L3J0 A0A1L8DEQ3 A0A1L8DEI6 A0A194QI00 A0A2A4J5Q3 A0A2H1VFS1 A0A182KSD5 A0A194PT22 A0A194QC91 A0A2W1BRJ1 A0A2A4JC88 A0A0N1PI45 A0A0L7LKI8 A0A194RIS2 A0A212F2Q3 A0A182MYV4 A0A2A4K5R4 A0A2A4K5P1 A0A182RFF8 A0A195E462 B0XDE9 A0A0K8VUC5 A0A1Y1KD84 A0A0K8UFN6
A0A2A4K5Q3 A0A2W1BRL6 A0A3S2TCP3 A0A212F123 A0A194PR44 A0A212F124 A0A194RF38 A0A0L7KZ57 A0A194RJF2 H9JI52 A0A2A4J6J0 A0A0L7KWH4 A0A194RGD2 A0A212F137 A0A194PJI6 A0A194RF43 A0A2A4K5Q2 A0A3S2L9U7 A0A212EJ24 A0A212EMB5 A0A2W1BMH3 A0A0L7KTQ3 A0A194RE10 A0A194PT27 A0A2H1VQB3 A0A194PSH3 A0A2A4K663 A0A2H1VCJ6 A0A2H1VQC6 A0A2W1BSR9 A0A2A4JIA3 A0A2H1VAT9 A0A2W1BHB5 A0A2A4JH68 A0A2W1BLR2 A0A3S2LQC2 A0A2H1VSF4 A0A0L7K3V5 A0A2A4K6L7 A0A0L7KTW1 A0A2W1BMV0 A0A2H1V9B3 H9JI48 A0A3S2NA23 A0A0L7KVH9 A0A0L7K4R7 A0A194QDP3 A0A212FPC5 A0A3S2NDN7 A0A182SYI8 B0XDF1 A0A2J7RPZ7 Q7Q0C3 Q16WK7 A0A2A4K6M9 B0XDF0 A0A2J7RPY7 A0A182LZQ5 A0A2J7RPY0 A0A067RSY2 A0A3S2M6B8 A0A212EJ20 A0A2H1VA76 A0A182Y1J3 A0A3S2NIQ2 A0A182QUL1 A0A182GYP2 A0A3S2L3J0 A0A1L8DEQ3 A0A1L8DEI6 A0A194QI00 A0A2A4J5Q3 A0A2H1VFS1 A0A182KSD5 A0A194PT22 A0A194QC91 A0A2W1BRJ1 A0A2A4JC88 A0A0N1PI45 A0A0L7LKI8 A0A194RIS2 A0A212F2Q3 A0A182MYV4 A0A2A4K5R4 A0A2A4K5P1 A0A182RFF8 A0A195E462 B0XDE9 A0A0K8VUC5 A0A1Y1KD84 A0A0K8UFN6
PDB
4LDS
E-value=8.70234e-11,
Score=162
Ontologies
GO
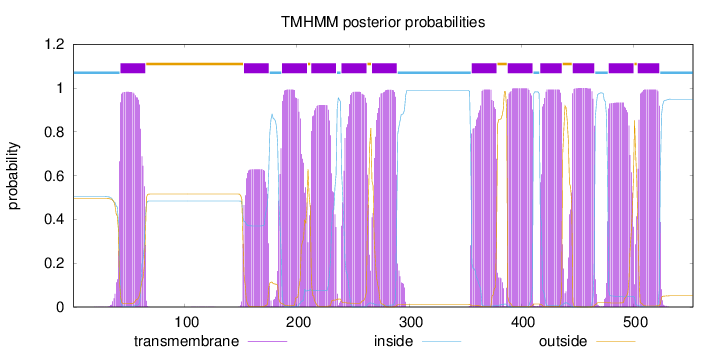
Topology
Length:
553
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
247.47897
Exp number, first 60 AAs:
18.75908
Total prob of N-in:
0.50424
POSSIBLE N-term signal
sequence
inside
1 - 42
TMhelix
43 - 65
outside
66 - 152
TMhelix
153 - 175
inside
176 - 186
TMhelix
187 - 209
outside
210 - 212
TMhelix
213 - 235
inside
236 - 239
TMhelix
240 - 262
outside
263 - 266
TMhelix
267 - 289
inside
290 - 355
TMhelix
356 - 378
outside
379 - 387
TMhelix
388 - 410
inside
411 - 416
TMhelix
417 - 436
outside
437 - 445
TMhelix
446 - 465
inside
466 - 477
TMhelix
478 - 500
outside
501 - 503
TMhelix
504 - 523
inside
524 - 553
Population Genetic Test Statistics
Pi
198.271712
Theta
179.777054
Tajima's D
0.777413
CLR
11.876316
CSRT
0.592320383980801
Interpretation
Uncertain
