Gene
KWMTBOMO13503
Pre Gene Modal
BGIBMGA009199
Annotation
PREDICTED:_organic_cation_transporter_protein-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.761
Sequence
CDS
ATGACCGCAGTGGCAGCGGGAGTAGCAGGAGTCGCCAGAGCATTCTCCAACTCATACGTCCTCTACGTAGTGCTTGAGTTCGTGGAGGCCACTGTTGGAGCCGGAGCTTATAGTACTGGGTTCATTCTTGCTCTGGAAATGGTGGGAGTAAACCGTAGAGTTCTCGGTGGGAATATCATATCAAGCACTTTCGCACTAGGACAAGTGGTGGTGGCCCTTACAGCGTGGGCGTTCCCGTACTGGCGTACTTTGACCTTGATTATCTACGCTCCATCAATACTATTCATCCTATACATCTTTGTCATAGAAGAGAGTGTCAGATGGCTGATAAGTAAAGGAAGGAAGAAGGAAGCTGCCAAAATCATTTTCAAAGCCGCAGCAATTAACAATAAGAAGTTGTCGCCAGAAACTATCAAAATACTTAATGATGAAAGCGTTGTAGAGGAGGAAACTAAATCCCCAGTTAAAGAACTACCGAAAAAATCCCTAGCAGTGCAGGTTTTGAGGTCTAAAACTATAATGACCCGGCTTTGTGTTTGTTGTTTTTGGTGGATAACGCTTACCTTCGTATACTACGGGCTGTCGATCAACTCCGTGTCTTTGGCTGGGAACAGCTACGTCAATTACATTTTGACTTCACTAGTGGAGATCCCCGGATACTGTCTCAGTGTGCTGACGATGGACAGGTTCGGGAGGAAAGCCTCGATCATGACTGCCTTTATTGTGTGTGGCCTGTCGCTCGTCGGATTGCCGTTTGTACCCGAGAACATCATTTGGCTCCAAACGACGCTCAACATGATCGGAAAGTTGTGCATCAGTATGGCGTTCAGCAGTATATATATATACACATCAGAGCTGTTCCCCACTGAAGCGAGACACACTCTCCTCGGCGCCTGCTCCATGATTGGACGGATAGGGTCCTTAGTTGCGCCGCAGACTCCTTTACTGATGACATACATGGAGTCTCTTCCGTACCTGATTTTCGGCATAATGGCGTGTACATCTGGTCTACTGATGCTTCTAACACCAGAAACCCTGAGGGTAAAGCTGCCAGACACCATAGAACAGGCAGAGAACATCAGCAAAAAGCCACAGAACGGAACTCTTAATGGTGTCGTAACCTCAACTTGA
Protein
MTAVAAGVAGVARAFSNSYVLYVVLEFVEATVGAGAYSTGFILALEMVGVNRRVLGGNIISSTFALGQVVVALTAWAFPYWRTLTLIIYAPSILFILYIFVIEESVRWLISKGRKKEAAKIIFKAAAINNKKLSPETIKILNDESVVEEETKSPVKELPKKSLAVQVLRSKTIMTRLCVCCFWWITLTFVYYGLSINSVSLAGNSYVNYILTSLVEIPGYCLSVLTMDRFGRKASIMTAFIVCGLSLVGLPFVPENIIWLQTTLNMIGKLCISMAFSSIYIYTSELFPTEARHTLLGACSMIGRIGSLVAPQTPLLMTYMESLPYLIFGIMACTSGLLMLLTPETLRVKLPDTIEQAENISKKPQNGTLNGVVTST
Summary
Uniprot
H9JI51
A0A0L7KUG4
A0A2H1V9B3
A0A2A4K6M9
A0A3S2NA23
A0A194PSH8
+ More
A0A194RE04 A0A212F848 A0A2A4K5P1 A0A2H1V9G0 A0A2W1BME8 A0A212EJ20 A0A0L7LSA4 A0A194RE15 A0A2H1VA76 A0A2A4K7E5 A0A3S2NIQ2 A0A3S2M6B8 A0A2A4K7E3 A0A2A4K5R4 A0A2W1BRJ1 A0A194PT22 A0A195E462 A0A2W1BPN7 A0A026W5E8 A0A3L8D7U5 A0A212EJ24 A0A158P2C5 A0A067RGL9 A0A154PCE9 H9IVN2 F4WUN7 E2BPX2 A0A2J7RPY7 A0A2J7RPY0 A0A0L7RIG1 A0A2H1V1L3 A0A194RF43 V9ICM7 A0A2A3E3H4 A0A088AAF9 E2A7K4 A0A2A4K5Q2 A0A195DF58 A0A2W1BMH3 A0A0T6AWJ5 A0A158NQD1 A0A194RGD2 A0A212FPC5 A0A2A4JBT3 A0A0L7KTQ3 A0A194PT27 A0A194RE10 A0A2J7RPY2 A0A195FIT4 A0A2H1VCJ6 A0A2H1VQC6 A0A2A4K663 A0A151WZG1 A0A195D353 A0A195CVD4 H9IWG7 A0A151WZE2 A0A067RSY2 A0A0V0GBF3 A0A2W1BSR9 A0A2A4JIA3 A0A3S2L3J0 A0A2A4JC88 A0A151WY09 A0A2J7RPY5 A0A2H1VSF4 A0A194PJI6 A0A195BUZ4 A0A2H1VFS1 A0A0L7LKI8 A0A2J7RPY4 A0A0L7K3V5 A0A3S2L9U7 A0A212F2Q3 A0A2A4J455 A0A2A4J6J0 A0A2J7RPZ7 A0A194REW9 F4WUN6 A0A2H1VQB3 A0A2A4J5Q3 A0A2W1BLR2 A0A3S2NDN7 A0A3S2LQC2 A0A1L8DEQ3 A0A2W1BHB5 A0A1L8DEI6 A0A195E542 A0A195ETW6 A0A084WCB2 A0A2A4JH68 A0A194QC06 A0A2M3ZIE3 A0A2A4K6L7
A0A194RE04 A0A212F848 A0A2A4K5P1 A0A2H1V9G0 A0A2W1BME8 A0A212EJ20 A0A0L7LSA4 A0A194RE15 A0A2H1VA76 A0A2A4K7E5 A0A3S2NIQ2 A0A3S2M6B8 A0A2A4K7E3 A0A2A4K5R4 A0A2W1BRJ1 A0A194PT22 A0A195E462 A0A2W1BPN7 A0A026W5E8 A0A3L8D7U5 A0A212EJ24 A0A158P2C5 A0A067RGL9 A0A154PCE9 H9IVN2 F4WUN7 E2BPX2 A0A2J7RPY7 A0A2J7RPY0 A0A0L7RIG1 A0A2H1V1L3 A0A194RF43 V9ICM7 A0A2A3E3H4 A0A088AAF9 E2A7K4 A0A2A4K5Q2 A0A195DF58 A0A2W1BMH3 A0A0T6AWJ5 A0A158NQD1 A0A194RGD2 A0A212FPC5 A0A2A4JBT3 A0A0L7KTQ3 A0A194PT27 A0A194RE10 A0A2J7RPY2 A0A195FIT4 A0A2H1VCJ6 A0A2H1VQC6 A0A2A4K663 A0A151WZG1 A0A195D353 A0A195CVD4 H9IWG7 A0A151WZE2 A0A067RSY2 A0A0V0GBF3 A0A2W1BSR9 A0A2A4JIA3 A0A3S2L3J0 A0A2A4JC88 A0A151WY09 A0A2J7RPY5 A0A2H1VSF4 A0A194PJI6 A0A195BUZ4 A0A2H1VFS1 A0A0L7LKI8 A0A2J7RPY4 A0A0L7K3V5 A0A3S2L9U7 A0A212F2Q3 A0A2A4J455 A0A2A4J6J0 A0A2J7RPZ7 A0A194REW9 F4WUN6 A0A2H1VQB3 A0A2A4J5Q3 A0A2W1BLR2 A0A3S2NDN7 A0A3S2LQC2 A0A1L8DEQ3 A0A2W1BHB5 A0A1L8DEI6 A0A195E542 A0A195ETW6 A0A084WCB2 A0A2A4JH68 A0A194QC06 A0A2M3ZIE3 A0A2A4K6L7
Pubmed
EMBL
BABH01041111
JTDY01005496
KOB66937.1
ODYU01001366
SOQ37440.1
NWSH01000100
+ More
PCG79564.1 RSAL01000360 RVE42197.1 KQ459595 KPI95923.1 KQ460313 KPJ16063.1 AGBW02009797 OWR49901.1 PCG79565.1 SOQ37441.1 KZ149964 PZC76252.1 AGBW02014542 OWR41475.1 JTDY01000245 KOB78076.1 KPJ16073.1 ODYU01001220 SOQ37134.1 PCG79572.1 RSAL01000974 RVE41008.1 RSAL01000027 RVE51971.1 PCG79570.1 PCG79571.1 PZC76264.1 KPI95914.1 KQ979657 KYN19975.1 PZC76251.1 KK107403 EZA51300.1 QOIP01000011 RLU16525.1 OWR41479.1 ADTU01007112 KK852482 KDR22927.1 KQ434870 KZC09576.1 BABH01039623 GL888375 EGI62034.1 GL449663 EFN82259.1 NEVH01001354 PNF42889.1 PNF42891.1 KQ414585 KOC70516.1 ODYU01000079 SOQ34232.1 KPJ16069.1 JR038305 AEY58206.1 KZ288444 PBC25689.1 GL437348 EFN70576.1 PCG79557.1 KQ980905 KYN11533.1 PZC76259.1 LJIG01022634 KRT79523.1 ADTU01001734 KQ460205 KPJ16878.1 AGBW02003000 OWR55596.1 NWSH01002161 PCG69014.1 JTDY01005909 KOB66490.1 KPI95919.1 KPJ16068.1 PNF42895.1 KQ981523 KYN40281.1 ODYU01001566 SOQ37954.1 ODYU01003788 SOQ42986.1 PCG79559.1 KQ982639 KYQ53259.1 KQ976885 KYN07327.1 KQ977259 KYN04595.1 BABH01006647 KYQ53258.1 KDR22929.1 GECL01000840 JAP05284.1 PZC76257.1 NWSH01001466 PCG71160.1 RSAL01000186 RVE44802.1 PCG69013.1 KQ982662 KYQ52637.1 PNF42894.1 ODYU01004183 SOQ43775.1 KQ459606 KPI91250.1 KQ976408 KYM91167.1 ODYU01002328 SOQ39679.1 JTDY01000842 KOB75696.1 PNF42897.1 JTDY01011376 KOB56709.1 RVE42203.1 AGBW02010692 OWR48028.1 NWSH01003151 PCG66855.1 NWSH01002815 PCG67489.1 PNF42898.1 KQ460325 KPJ15840.1 EGI62033.1 SOQ42987.1 PCG66854.1 KZ150061 PZC74227.1 RVE44804.1 RVE42202.1 GFDF01009227 JAV04857.1 PZC74228.1 GFDF01009228 JAV04856.1 KYN19974.1 KQ981979 KYN31324.1 ATLV01022651 KE525335 KFB47856.1 PCG71159.1 KQ459193 KPJ03083.1 GGFM01007521 MBW28272.1 PCG79558.1
PCG79564.1 RSAL01000360 RVE42197.1 KQ459595 KPI95923.1 KQ460313 KPJ16063.1 AGBW02009797 OWR49901.1 PCG79565.1 SOQ37441.1 KZ149964 PZC76252.1 AGBW02014542 OWR41475.1 JTDY01000245 KOB78076.1 KPJ16073.1 ODYU01001220 SOQ37134.1 PCG79572.1 RSAL01000974 RVE41008.1 RSAL01000027 RVE51971.1 PCG79570.1 PCG79571.1 PZC76264.1 KPI95914.1 KQ979657 KYN19975.1 PZC76251.1 KK107403 EZA51300.1 QOIP01000011 RLU16525.1 OWR41479.1 ADTU01007112 KK852482 KDR22927.1 KQ434870 KZC09576.1 BABH01039623 GL888375 EGI62034.1 GL449663 EFN82259.1 NEVH01001354 PNF42889.1 PNF42891.1 KQ414585 KOC70516.1 ODYU01000079 SOQ34232.1 KPJ16069.1 JR038305 AEY58206.1 KZ288444 PBC25689.1 GL437348 EFN70576.1 PCG79557.1 KQ980905 KYN11533.1 PZC76259.1 LJIG01022634 KRT79523.1 ADTU01001734 KQ460205 KPJ16878.1 AGBW02003000 OWR55596.1 NWSH01002161 PCG69014.1 JTDY01005909 KOB66490.1 KPI95919.1 KPJ16068.1 PNF42895.1 KQ981523 KYN40281.1 ODYU01001566 SOQ37954.1 ODYU01003788 SOQ42986.1 PCG79559.1 KQ982639 KYQ53259.1 KQ976885 KYN07327.1 KQ977259 KYN04595.1 BABH01006647 KYQ53258.1 KDR22929.1 GECL01000840 JAP05284.1 PZC76257.1 NWSH01001466 PCG71160.1 RSAL01000186 RVE44802.1 PCG69013.1 KQ982662 KYQ52637.1 PNF42894.1 ODYU01004183 SOQ43775.1 KQ459606 KPI91250.1 KQ976408 KYM91167.1 ODYU01002328 SOQ39679.1 JTDY01000842 KOB75696.1 PNF42897.1 JTDY01011376 KOB56709.1 RVE42203.1 AGBW02010692 OWR48028.1 NWSH01003151 PCG66855.1 NWSH01002815 PCG67489.1 PNF42898.1 KQ460325 KPJ15840.1 EGI62033.1 SOQ42987.1 PCG66854.1 KZ150061 PZC74227.1 RVE44804.1 RVE42202.1 GFDF01009227 JAV04857.1 PZC74228.1 GFDF01009228 JAV04856.1 KYN19974.1 KQ981979 KYN31324.1 ATLV01022651 KE525335 KFB47856.1 PCG71159.1 KQ459193 KPJ03083.1 GGFM01007521 MBW28272.1 PCG79558.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000283053
UP000053268
UP000053240
+ More
UP000007151 UP000078492 UP000053097 UP000279307 UP000005205 UP000027135 UP000076502 UP000007755 UP000008237 UP000235965 UP000053825 UP000242457 UP000005203 UP000000311 UP000078541 UP000075809 UP000078542 UP000078540 UP000030765
UP000007151 UP000078492 UP000053097 UP000279307 UP000005205 UP000027135 UP000076502 UP000007755 UP000008237 UP000235965 UP000053825 UP000242457 UP000005203 UP000000311 UP000078541 UP000075809 UP000078542 UP000078540 UP000030765
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JI51
A0A0L7KUG4
A0A2H1V9B3
A0A2A4K6M9
A0A3S2NA23
A0A194PSH8
+ More
A0A194RE04 A0A212F848 A0A2A4K5P1 A0A2H1V9G0 A0A2W1BME8 A0A212EJ20 A0A0L7LSA4 A0A194RE15 A0A2H1VA76 A0A2A4K7E5 A0A3S2NIQ2 A0A3S2M6B8 A0A2A4K7E3 A0A2A4K5R4 A0A2W1BRJ1 A0A194PT22 A0A195E462 A0A2W1BPN7 A0A026W5E8 A0A3L8D7U5 A0A212EJ24 A0A158P2C5 A0A067RGL9 A0A154PCE9 H9IVN2 F4WUN7 E2BPX2 A0A2J7RPY7 A0A2J7RPY0 A0A0L7RIG1 A0A2H1V1L3 A0A194RF43 V9ICM7 A0A2A3E3H4 A0A088AAF9 E2A7K4 A0A2A4K5Q2 A0A195DF58 A0A2W1BMH3 A0A0T6AWJ5 A0A158NQD1 A0A194RGD2 A0A212FPC5 A0A2A4JBT3 A0A0L7KTQ3 A0A194PT27 A0A194RE10 A0A2J7RPY2 A0A195FIT4 A0A2H1VCJ6 A0A2H1VQC6 A0A2A4K663 A0A151WZG1 A0A195D353 A0A195CVD4 H9IWG7 A0A151WZE2 A0A067RSY2 A0A0V0GBF3 A0A2W1BSR9 A0A2A4JIA3 A0A3S2L3J0 A0A2A4JC88 A0A151WY09 A0A2J7RPY5 A0A2H1VSF4 A0A194PJI6 A0A195BUZ4 A0A2H1VFS1 A0A0L7LKI8 A0A2J7RPY4 A0A0L7K3V5 A0A3S2L9U7 A0A212F2Q3 A0A2A4J455 A0A2A4J6J0 A0A2J7RPZ7 A0A194REW9 F4WUN6 A0A2H1VQB3 A0A2A4J5Q3 A0A2W1BLR2 A0A3S2NDN7 A0A3S2LQC2 A0A1L8DEQ3 A0A2W1BHB5 A0A1L8DEI6 A0A195E542 A0A195ETW6 A0A084WCB2 A0A2A4JH68 A0A194QC06 A0A2M3ZIE3 A0A2A4K6L7
A0A194RE04 A0A212F848 A0A2A4K5P1 A0A2H1V9G0 A0A2W1BME8 A0A212EJ20 A0A0L7LSA4 A0A194RE15 A0A2H1VA76 A0A2A4K7E5 A0A3S2NIQ2 A0A3S2M6B8 A0A2A4K7E3 A0A2A4K5R4 A0A2W1BRJ1 A0A194PT22 A0A195E462 A0A2W1BPN7 A0A026W5E8 A0A3L8D7U5 A0A212EJ24 A0A158P2C5 A0A067RGL9 A0A154PCE9 H9IVN2 F4WUN7 E2BPX2 A0A2J7RPY7 A0A2J7RPY0 A0A0L7RIG1 A0A2H1V1L3 A0A194RF43 V9ICM7 A0A2A3E3H4 A0A088AAF9 E2A7K4 A0A2A4K5Q2 A0A195DF58 A0A2W1BMH3 A0A0T6AWJ5 A0A158NQD1 A0A194RGD2 A0A212FPC5 A0A2A4JBT3 A0A0L7KTQ3 A0A194PT27 A0A194RE10 A0A2J7RPY2 A0A195FIT4 A0A2H1VCJ6 A0A2H1VQC6 A0A2A4K663 A0A151WZG1 A0A195D353 A0A195CVD4 H9IWG7 A0A151WZE2 A0A067RSY2 A0A0V0GBF3 A0A2W1BSR9 A0A2A4JIA3 A0A3S2L3J0 A0A2A4JC88 A0A151WY09 A0A2J7RPY5 A0A2H1VSF4 A0A194PJI6 A0A195BUZ4 A0A2H1VFS1 A0A0L7LKI8 A0A2J7RPY4 A0A0L7K3V5 A0A3S2L9U7 A0A212F2Q3 A0A2A4J455 A0A2A4J6J0 A0A2J7RPZ7 A0A194REW9 F4WUN6 A0A2H1VQB3 A0A2A4J5Q3 A0A2W1BLR2 A0A3S2NDN7 A0A3S2LQC2 A0A1L8DEQ3 A0A2W1BHB5 A0A1L8DEI6 A0A195E542 A0A195ETW6 A0A084WCB2 A0A2A4JH68 A0A194QC06 A0A2M3ZIE3 A0A2A4K6L7
PDB
4J05
E-value=0.000405573,
Score=103
Ontologies
GO
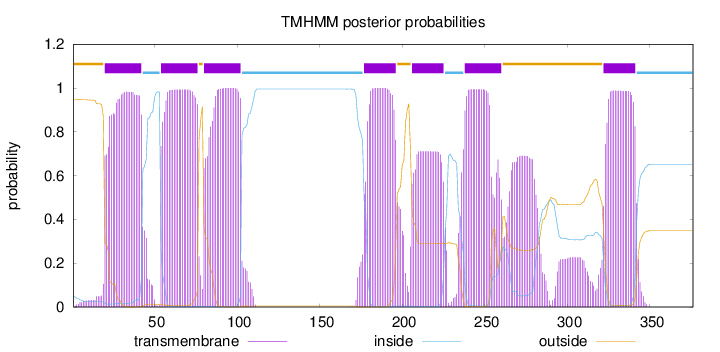
Topology
Length:
376
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
165.20104
Exp number, first 60 AAs:
29.57834
Total prob of N-in:
0.05074
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 42
inside
43 - 53
TMhelix
54 - 76
outside
77 - 79
TMhelix
80 - 102
inside
103 - 176
TMhelix
177 - 196
outside
197 - 205
TMhelix
206 - 225
inside
226 - 237
TMhelix
238 - 260
outside
261 - 321
TMhelix
322 - 341
inside
342 - 376
Population Genetic Test Statistics
Pi
273.347362
Theta
211.751471
Tajima's D
0.913034
CLR
0.036925
CSRT
0.631468426578671
Interpretation
Possibly Positive selection
