Gene
KWMTBOMO13498
Pre Gene Modal
BGIBMGA009185
Annotation
PREDICTED:_organic_cation_transporter_protein-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.91
Sequence
CDS
ATGGTGGCGAGTGCGCACGCGTCGCGCGACCTCGCCGCCGAAGCCCTAGAGGACGCTCTGGGGCGACTCGGGCCCTTCGGCCTCTACCAGCGATATGTCGCGATAATGCTGTGTATCCCCAACCTGCTCGCGGCCATGTATACCCTTAATTATGTGTTCGTTGCGGACCAGGTGCCCTTTCGATGTGTGGTCCCGGAATGCGAAGGTAACTACACCCAGTTCCACAACCCTGCAGCACAGGCCCTCCTCCCGGCGGACTCCCGCTGCGACCGATACTTACCACTGCAGGACGCAGGGACAGTCTCCTGTCAGCGAGAAGACTTCCATCAGAATGACACTAAGGCTTGCAATCATTTTGTATATGAGAGCATGTCTACTACTTTTGCTGAGTTTAATTTGGCGTGTCGCGAGTGGATGCGAACCCTGGTCGGCACCGTCCGTAACGCGGCGCTGCCCCTCGCGCTCCTGCTCACAGGATACATCTCCGACACATATGGTCGCCGCACGGCTTTCTGTATATTCTCAGCCTTCGCGGCAGTGCTCGGCATTGTCAAGGGCCTCTCCATCAACTACCAGATGTATGTCGCCATGGAGTTCTTTGAGGCAGCCCTCGGCTATGGCTTTAATAGCGCTGGATATGTCATGATGGTGGAGCTGGCTCGACCATCGCTCCGAGCTTCGTTCGCGTGCGCCACGGGAGTCGCCTACGGTCTGGGCGGGATGCTGTTCGCTCTTATAGCGTGGAAGATACAGTACTGGCGGCATCTGCTGTACACCGTACACTCCTTCGCGCTGCTGCTGCCACTCTACTGGCTTCTGGTAGACGAGAGTCCGCGCTGGCTACACGCAAAGGGCAGAACTGAGGAAACCGTGGACATCATCAAGAAGGCTGCCAGGTGGAACAAGGTCCAAATAAGTGAAGAAATTCTCTTCAAGTTGAGCGACAAGGATTCCATGAAGTCAGATTCAGACTCAACCAGCTCCAACCCTTGGCTGCGGCTCCTGAGGTCCCGCGTGCTAGTGGGACGCTTCGCGATCTGCTCCTGGTGCTGGGTGGCGGTCGCTTTCGTCTACTACGGACTCACTATAAACTCGGTCTCGCTCTCGGGAGATAAGTACGCGAACTTTGCTCTGAATATGTCGATGGAGATAGTGGCGTCGCTACTGATCATGATGGCGCTGGAGAGGTTCGGCCGGAAGTGGAGCATCTTCGTCGCGTTCCTGGTCTGCGGCGTCGCTTGTGTCACACCTTACTTCACTTCGCACGCGGGCGCGGGGCTGGGGATGTTCTTCGTGGGCAAGCTGGCGATAACATTCGCCTTCAACTCCATGTACGTGTTCACGGCGGAGCTGTTCCCGACCAGCGTCCGCAGCTCCGCGCTGGCGGCCTGCTCGCTCGTGGGCAGGGTCGGCTCTATACTGGCGCCGCAGACTCCCTTACTTAGTATATACATCCAAGCTTTGTTATACGGCGCGTTCTCGTGTCTGGCGGCGCTGGCGGTGCTGATGGTGCCGGAGACGAGGCGGGCCATGCTGCCGCGTGACGTCACTGACGCTGAACACATCCGGGGCTCCTCTCCGAGCCCCGCCCCCGCGCCCGCTCCCGCCGACACCCCCCGCACCGTGGTGCTCTCCGCGGACACGTGA
Protein
MVASAHASRDLAAEALEDALGRLGPFGLYQRYVAIMLCIPNLLAAMYTLNYVFVADQVPFRCVVPECEGNYTQFHNPAAQALLPADSRCDRYLPLQDAGTVSCQREDFHQNDTKACNHFVYESMSTTFAEFNLACREWMRTLVGTVRNAALPLALLLTGYISDTYGRRTAFCIFSAFAAVLGIVKGLSINYQMYVAMEFFEAALGYGFNSAGYVMMVELARPSLRASFACATGVAYGLGGMLFALIAWKIQYWRHLLYTVHSFALLLPLYWLLVDESPRWLHAKGRTEETVDIIKKAARWNKVQISEEILFKLSDKDSMKSDSDSTSSNPWLRLLRSRVLVGRFAICSWCWVAVAFVYYGLTINSVSLSGDKYANFALNMSMEIVASLLIMMALERFGRKWSIFVAFLVCGVACVTPYFTSHAGAGLGMFFVGKLAITFAFNSMYVFTAELFPTSVRSSALAACSLVGRVGSILAPQTPLLSIYIQALLYGAFSCLAALAVLMVPETRRAMLPRDVTDAEHIRGSSPSPAPAPAPADTPRTVVLSADT
Summary
Uniprot
H9JI37
A0A2H1VSB0
A0A212EIZ7
A0A2A4K5Q8
A0A0L7L0F4
A0A194PR33
+ More
A0A194RJG1 A0A2A4K6M6 A0A3S2NA23 A0A2A4K5R4 A0A2H1VA76 A0A3S2M6B8 A0A3S2NIQ2 A0A2W1BRJ1 A0A2A4K7E3 A0A212EJ20 A0A194PT22 A0A2H1V9B3 H9IVN2 A0A0L7KUG4 A0A194PSH8 A0A2A4K6M9 A0A3L8D7U5 A0A212FPC5 A0A026W5E8 A0A182SYI8 A0A2H1V9G0 A0A1L8DEQ3 A0A154PCE9 A0A1L8DEI6 A0A3S2L3J0 E2A7K4 A0A194QDP3 A0A195DF58 A0A0N1PI45 A0A194QI00 A0A212F848 A0A2A4K7D4 A0A1B0DMI4 A0A2W1BUD3 V9ICM7 A0A2A3E3H4 U5EWC3 A0A3S2NDN7 A0A2H1VFS1 A0A088AAF9 A0A212F2Q3 A0A1I8MQZ5 A0A212F158 A0A2A4JC88 A0A0L7KWH4 F4WUN7 A0A1B0CF89 A0A0L7RIG1 A0A194QC91 A0A151WZG1 A0A195CVD4 A0A2J7RPY5 A0A182FXJ6 A0A194RIS2 A0A2A4K4I8 A0A158P2C5 B3NJ17 A0A1B0D118 W8BY90 A0A084WCB2 A0A1L8DED6 B4IUN3 A0A0K8VUC5 A0A2W1BM98 B4J0Q6 A0A182WZR9 A0A182VDY0 A0A0K8VEY5 A0A0A1X788 A0A034VVY9 A0A0K8UAY5 A0A0K8UFN6 A0A0L0CC92 Q9VNW8 A0A0L0C9F0 A0A182KSC8 A0A0J9URD0 B3MAJ1 A0A195D353 A0A0A1XJ72 A0A182JXE2 B4IAX7 A0A1L8DER5 A0A0K8VF65 A0A182TM67 A0A195FIT4 A0A212EJ24 A0A3B0JLU9 B4QKP9 A0A034WIX2 A0A0Q9WR15 Q9VNX2 B4IAX2
A0A194RJG1 A0A2A4K6M6 A0A3S2NA23 A0A2A4K5R4 A0A2H1VA76 A0A3S2M6B8 A0A3S2NIQ2 A0A2W1BRJ1 A0A2A4K7E3 A0A212EJ20 A0A194PT22 A0A2H1V9B3 H9IVN2 A0A0L7KUG4 A0A194PSH8 A0A2A4K6M9 A0A3L8D7U5 A0A212FPC5 A0A026W5E8 A0A182SYI8 A0A2H1V9G0 A0A1L8DEQ3 A0A154PCE9 A0A1L8DEI6 A0A3S2L3J0 E2A7K4 A0A194QDP3 A0A195DF58 A0A0N1PI45 A0A194QI00 A0A212F848 A0A2A4K7D4 A0A1B0DMI4 A0A2W1BUD3 V9ICM7 A0A2A3E3H4 U5EWC3 A0A3S2NDN7 A0A2H1VFS1 A0A088AAF9 A0A212F2Q3 A0A1I8MQZ5 A0A212F158 A0A2A4JC88 A0A0L7KWH4 F4WUN7 A0A1B0CF89 A0A0L7RIG1 A0A194QC91 A0A151WZG1 A0A195CVD4 A0A2J7RPY5 A0A182FXJ6 A0A194RIS2 A0A2A4K4I8 A0A158P2C5 B3NJ17 A0A1B0D118 W8BY90 A0A084WCB2 A0A1L8DED6 B4IUN3 A0A0K8VUC5 A0A2W1BM98 B4J0Q6 A0A182WZR9 A0A182VDY0 A0A0K8VEY5 A0A0A1X788 A0A034VVY9 A0A0K8UAY5 A0A0K8UFN6 A0A0L0CC92 Q9VNW8 A0A0L0C9F0 A0A182KSC8 A0A0J9URD0 B3MAJ1 A0A195D353 A0A0A1XJ72 A0A182JXE2 B4IAX7 A0A1L8DER5 A0A0K8VF65 A0A182TM67 A0A195FIT4 A0A212EJ24 A0A3B0JLU9 B4QKP9 A0A034WIX2 A0A0Q9WR15 Q9VNX2 B4IAX2
Pubmed
EMBL
BABH01041187
BABH01041188
ODYU01004125
SOQ43666.1
AGBW02014543
OWR41473.1
+ More
NWSH01000100 PCG79567.1 JTDY01003949 KOB68791.1 KQ459595 KPI95911.1 KQ460313 KPJ16076.1 PCG79568.1 RSAL01000360 RVE42197.1 PCG79571.1 ODYU01001220 SOQ37134.1 RSAL01000027 RVE51971.1 RSAL01000974 RVE41008.1 KZ149964 PZC76264.1 PCG79570.1 AGBW02014542 OWR41475.1 KPI95914.1 ODYU01001366 SOQ37440.1 BABH01039623 JTDY01005496 KOB66937.1 KPI95923.1 PCG79564.1 QOIP01000011 RLU16525.1 AGBW02003000 OWR55596.1 KK107403 EZA51300.1 SOQ37441.1 GFDF01009227 JAV04857.1 KQ434870 KZC09576.1 GFDF01009228 JAV04856.1 RSAL01000186 RVE44802.1 GL437348 EFN70576.1 KQ459193 KPJ03085.1 KQ980905 KYN11533.1 KQ459762 KPJ20130.1 KPJ03081.1 AGBW02009797 OWR49901.1 PCG79560.1 AJVK01006968 PZC76256.1 JR038305 AEY58206.1 KZ288444 PBC25689.1 GANO01000632 JAB59239.1 RVE44804.1 ODYU01002328 SOQ39679.1 AGBW02010692 OWR48028.1 AGBW02010973 OWR47451.1 NWSH01002161 PCG69013.1 JTDY01005018 KOB67470.1 GL888375 EGI62034.1 AJWK01009829 KQ414585 KOC70516.1 KPJ03082.1 KQ982639 KYQ53259.1 KQ977259 KYN04595.1 NEVH01001354 PNF42894.1 KQ460325 KPJ15841.1 NWSH01000168 PCG78824.1 ADTU01007112 CH954178 EDV52734.1 AJVK01021828 GAMC01012331 JAB94224.1 ATLV01022651 KE525335 KFB47856.1 GFDF01009262 JAV04822.1 CM000159 CH891970 EDW95499.1 EDX00097.1 GDHF01009833 JAI42481.1 KZ149965 PZC76202.1 CH916366 EDV97911.1 GDHF01014878 JAI37436.1 GBXI01007300 JAD06992.1 GAKP01013274 JAC45678.1 GDHF01028477 JAI23837.1 GDHF01026931 GDHF01013660 GDHF01009296 JAI25383.1 JAI38654.1 JAI43018.1 JRES01000704 KNC29064.1 AE014296 BT150232 AAF51797.3 AGU12793.1 KNC29063.1 CM002912 KMZ01136.1 CH902618 EDV41278.1 KQ976885 KYN07327.1 GBXI01008187 GBXI01003290 JAD06105.1 JAD11002.1 CH480826 EDW44440.1 GFDF01009156 JAV04928.1 GDHF01014725 JAI37589.1 KQ981523 KYN40281.1 OWR41479.1 OUUW01000002 SPP76400.1 CM000363 EDX11464.1 GAKP01005259 JAC53693.1 CH940647 KRF84583.1 BT001724 AAF51793.2 AAN71479.1 EDW44435.1
NWSH01000100 PCG79567.1 JTDY01003949 KOB68791.1 KQ459595 KPI95911.1 KQ460313 KPJ16076.1 PCG79568.1 RSAL01000360 RVE42197.1 PCG79571.1 ODYU01001220 SOQ37134.1 RSAL01000027 RVE51971.1 RSAL01000974 RVE41008.1 KZ149964 PZC76264.1 PCG79570.1 AGBW02014542 OWR41475.1 KPI95914.1 ODYU01001366 SOQ37440.1 BABH01039623 JTDY01005496 KOB66937.1 KPI95923.1 PCG79564.1 QOIP01000011 RLU16525.1 AGBW02003000 OWR55596.1 KK107403 EZA51300.1 SOQ37441.1 GFDF01009227 JAV04857.1 KQ434870 KZC09576.1 GFDF01009228 JAV04856.1 RSAL01000186 RVE44802.1 GL437348 EFN70576.1 KQ459193 KPJ03085.1 KQ980905 KYN11533.1 KQ459762 KPJ20130.1 KPJ03081.1 AGBW02009797 OWR49901.1 PCG79560.1 AJVK01006968 PZC76256.1 JR038305 AEY58206.1 KZ288444 PBC25689.1 GANO01000632 JAB59239.1 RVE44804.1 ODYU01002328 SOQ39679.1 AGBW02010692 OWR48028.1 AGBW02010973 OWR47451.1 NWSH01002161 PCG69013.1 JTDY01005018 KOB67470.1 GL888375 EGI62034.1 AJWK01009829 KQ414585 KOC70516.1 KPJ03082.1 KQ982639 KYQ53259.1 KQ977259 KYN04595.1 NEVH01001354 PNF42894.1 KQ460325 KPJ15841.1 NWSH01000168 PCG78824.1 ADTU01007112 CH954178 EDV52734.1 AJVK01021828 GAMC01012331 JAB94224.1 ATLV01022651 KE525335 KFB47856.1 GFDF01009262 JAV04822.1 CM000159 CH891970 EDW95499.1 EDX00097.1 GDHF01009833 JAI42481.1 KZ149965 PZC76202.1 CH916366 EDV97911.1 GDHF01014878 JAI37436.1 GBXI01007300 JAD06992.1 GAKP01013274 JAC45678.1 GDHF01028477 JAI23837.1 GDHF01026931 GDHF01013660 GDHF01009296 JAI25383.1 JAI38654.1 JAI43018.1 JRES01000704 KNC29064.1 AE014296 BT150232 AAF51797.3 AGU12793.1 KNC29063.1 CM002912 KMZ01136.1 CH902618 EDV41278.1 KQ976885 KYN07327.1 GBXI01008187 GBXI01003290 JAD06105.1 JAD11002.1 CH480826 EDW44440.1 GFDF01009156 JAV04928.1 GDHF01014725 JAI37589.1 KQ981523 KYN40281.1 OWR41479.1 OUUW01000002 SPP76400.1 CM000363 EDX11464.1 GAKP01005259 JAC53693.1 CH940647 KRF84583.1 BT001724 AAF51793.2 AAN71479.1 EDW44435.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000037510
UP000053268
UP000053240
+ More
UP000283053 UP000279307 UP000053097 UP000075901 UP000076502 UP000000311 UP000078492 UP000092462 UP000242457 UP000005203 UP000095301 UP000007755 UP000092461 UP000053825 UP000075809 UP000078542 UP000235965 UP000069272 UP000005205 UP000008711 UP000030765 UP000002282 UP000001070 UP000076407 UP000075903 UP000037069 UP000000803 UP000075882 UP000007801 UP000075881 UP000001292 UP000075902 UP000078541 UP000268350 UP000000304 UP000008792
UP000283053 UP000279307 UP000053097 UP000075901 UP000076502 UP000000311 UP000078492 UP000092462 UP000242457 UP000005203 UP000095301 UP000007755 UP000092461 UP000053825 UP000075809 UP000078542 UP000235965 UP000069272 UP000005205 UP000008711 UP000030765 UP000002282 UP000001070 UP000076407 UP000075903 UP000037069 UP000000803 UP000075882 UP000007801 UP000075881 UP000001292 UP000075902 UP000078541 UP000268350 UP000000304 UP000008792
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JI37
A0A2H1VSB0
A0A212EIZ7
A0A2A4K5Q8
A0A0L7L0F4
A0A194PR33
+ More
A0A194RJG1 A0A2A4K6M6 A0A3S2NA23 A0A2A4K5R4 A0A2H1VA76 A0A3S2M6B8 A0A3S2NIQ2 A0A2W1BRJ1 A0A2A4K7E3 A0A212EJ20 A0A194PT22 A0A2H1V9B3 H9IVN2 A0A0L7KUG4 A0A194PSH8 A0A2A4K6M9 A0A3L8D7U5 A0A212FPC5 A0A026W5E8 A0A182SYI8 A0A2H1V9G0 A0A1L8DEQ3 A0A154PCE9 A0A1L8DEI6 A0A3S2L3J0 E2A7K4 A0A194QDP3 A0A195DF58 A0A0N1PI45 A0A194QI00 A0A212F848 A0A2A4K7D4 A0A1B0DMI4 A0A2W1BUD3 V9ICM7 A0A2A3E3H4 U5EWC3 A0A3S2NDN7 A0A2H1VFS1 A0A088AAF9 A0A212F2Q3 A0A1I8MQZ5 A0A212F158 A0A2A4JC88 A0A0L7KWH4 F4WUN7 A0A1B0CF89 A0A0L7RIG1 A0A194QC91 A0A151WZG1 A0A195CVD4 A0A2J7RPY5 A0A182FXJ6 A0A194RIS2 A0A2A4K4I8 A0A158P2C5 B3NJ17 A0A1B0D118 W8BY90 A0A084WCB2 A0A1L8DED6 B4IUN3 A0A0K8VUC5 A0A2W1BM98 B4J0Q6 A0A182WZR9 A0A182VDY0 A0A0K8VEY5 A0A0A1X788 A0A034VVY9 A0A0K8UAY5 A0A0K8UFN6 A0A0L0CC92 Q9VNW8 A0A0L0C9F0 A0A182KSC8 A0A0J9URD0 B3MAJ1 A0A195D353 A0A0A1XJ72 A0A182JXE2 B4IAX7 A0A1L8DER5 A0A0K8VF65 A0A182TM67 A0A195FIT4 A0A212EJ24 A0A3B0JLU9 B4QKP9 A0A034WIX2 A0A0Q9WR15 Q9VNX2 B4IAX2
A0A194RJG1 A0A2A4K6M6 A0A3S2NA23 A0A2A4K5R4 A0A2H1VA76 A0A3S2M6B8 A0A3S2NIQ2 A0A2W1BRJ1 A0A2A4K7E3 A0A212EJ20 A0A194PT22 A0A2H1V9B3 H9IVN2 A0A0L7KUG4 A0A194PSH8 A0A2A4K6M9 A0A3L8D7U5 A0A212FPC5 A0A026W5E8 A0A182SYI8 A0A2H1V9G0 A0A1L8DEQ3 A0A154PCE9 A0A1L8DEI6 A0A3S2L3J0 E2A7K4 A0A194QDP3 A0A195DF58 A0A0N1PI45 A0A194QI00 A0A212F848 A0A2A4K7D4 A0A1B0DMI4 A0A2W1BUD3 V9ICM7 A0A2A3E3H4 U5EWC3 A0A3S2NDN7 A0A2H1VFS1 A0A088AAF9 A0A212F2Q3 A0A1I8MQZ5 A0A212F158 A0A2A4JC88 A0A0L7KWH4 F4WUN7 A0A1B0CF89 A0A0L7RIG1 A0A194QC91 A0A151WZG1 A0A195CVD4 A0A2J7RPY5 A0A182FXJ6 A0A194RIS2 A0A2A4K4I8 A0A158P2C5 B3NJ17 A0A1B0D118 W8BY90 A0A084WCB2 A0A1L8DED6 B4IUN3 A0A0K8VUC5 A0A2W1BM98 B4J0Q6 A0A182WZR9 A0A182VDY0 A0A0K8VEY5 A0A0A1X788 A0A034VVY9 A0A0K8UAY5 A0A0K8UFN6 A0A0L0CC92 Q9VNW8 A0A0L0C9F0 A0A182KSC8 A0A0J9URD0 B3MAJ1 A0A195D353 A0A0A1XJ72 A0A182JXE2 B4IAX7 A0A1L8DER5 A0A0K8VF65 A0A182TM67 A0A195FIT4 A0A212EJ24 A0A3B0JLU9 B4QKP9 A0A034WIX2 A0A0Q9WR15 Q9VNX2 B4IAX2
PDB
4J05
E-value=0.0377629,
Score=88
Ontologies
GO
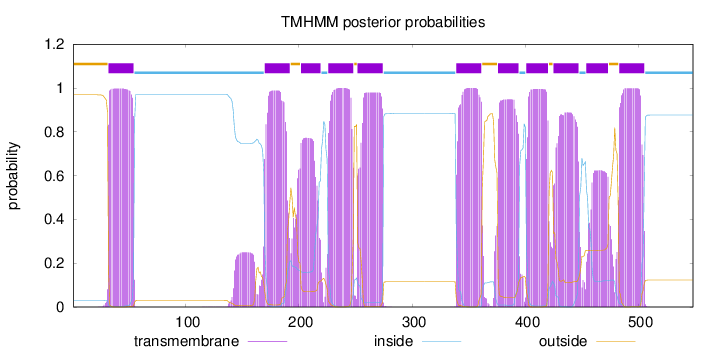
Topology
Length:
548
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
225.44384
Exp number, first 60 AAs:
22.79626
Total prob of N-in:
0.03107
POSSIBLE N-term signal
sequence
outside
1 - 31
TMhelix
32 - 54
inside
55 - 169
TMhelix
170 - 192
outside
193 - 201
TMhelix
202 - 219
inside
220 - 225
TMhelix
226 - 248
outside
249 - 251
TMhelix
252 - 274
inside
275 - 338
TMhelix
339 - 361
outside
362 - 375
TMhelix
376 - 394
inside
395 - 400
TMhelix
401 - 420
outside
421 - 424
TMhelix
425 - 447
inside
448 - 453
TMhelix
454 - 473
outside
474 - 482
TMhelix
483 - 505
inside
506 - 548
Population Genetic Test Statistics
Pi
22.818065
Theta
37.375562
Tajima's D
-1.775747
CLR
11.158521
CSRT
0.031748412579371
Interpretation
Uncertain
