Gene
KWMTBOMO13493 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009189
Annotation
Prefoldin_subunit_2_[Operophtera_brumata]
Full name
Prefoldin subunit 2
Location in the cell
Cytoplasmic Reliability : 2.146 Nuclear Reliability : 1.578
Sequence
CDS
ATGGAGTTAAACGAACATAAGATCGTAATTGAGACTTTGCGGGGAGTGGAGTTGACCCGTAAGTGTTTCCGGATGTTTGGGGGCGTGCTTGTCGAACGCACCGTAGCTGAGGTGTTGCCAGAGCTGATCAGCAATTATGAACAGCTACCTAAGGCCATCCAGTCCCTAGAAGACCAACTCACTCAGAAGGGAGAGGAGATCAATAAATACATTGAGGAACATGATATTCGCATCCAACGTGCTGATCGCACCATGCCTGAACCACCTCCAGAGCAATCCACCTCTACCAAGTCTAATGTGCTAGTTGCTAGCGGTTAA
Protein
MELNEHKIVIETLRGVELTRKCFRMFGGVLVERTVAEVLPELISNYEQLPKAIQSLEDQLTQKGEEINKYIEEHDIRIQRADRTMPEPPPEQSTSTKSNVLVASG
Summary
Description
Binds specifically to cytosolic chaperonin (c-CPN) and transfers target proteins to it. Binds to nascent polypeptide chain and promotes folding in an environment in which there are many competing pathways for nonnative proteins.
Binds specifically to cytosolic chaperonin (c-CPN) and transfers target proteins to it. Binds to nascent polypeptide chain and promotes folding in an environment in which there are many competing pathways for nonnative proteins (By similarity).
Binds specifically to cytosolic chaperonin (c-CPN) and transfers target proteins to it. Binds to nascent polypeptide chain and promotes folding in an environment in which there are many competing pathways for nonnative proteins (By similarity).
Subunit
Binds to c-Myc; interacts with its N-terminal domain (By similarity). Heterohexamer of two PFD-alpha type and four PFD-beta type subunits. Interacts with URI1; the interaction is phosphorylation-dependent and occurs in a growth-dependent manner.
Heterohexamer of two PFD-alpha type and four PFD-beta type subunits. Binds to c-Myc; interacts with its N-terminal domain. Interacts with URI1; the interaction is phosphorylation-dependent and occurs in a growth-dependent manner (By similarity).
Interacts with URI1; the interaction is phosphorylation-dependent and occurs in a growth-dependent manner (By similarity). Heterohexamer of two PFD-alpha type and four PFD-beta type subunits. Binds to c-Myc; interacts with its N-terminal domain.
Heterohexamer of two PFD-alpha type and four PFD-beta type subunits. Binds to c-Myc; interacts with its N-terminal domain. Interacts with URI1; the interaction is phosphorylation-dependent and occurs in a growth-dependent manner (By similarity).
Interacts with URI1; the interaction is phosphorylation-dependent and occurs in a growth-dependent manner (By similarity). Heterohexamer of two PFD-alpha type and four PFD-beta type subunits. Binds to c-Myc; interacts with its N-terminal domain.
Similarity
Belongs to the prefoldin subunit beta family.
Keywords
Chaperone
Complete proteome
Cytoplasm
Mitochondrion
Nucleus
Reference proteome
Feature
chain Prefoldin subunit 2
Uniprot
H9JI41
A0A2H1VM24
A0A2W1B4U1
A0A2A4J137
A0A1E1WBX3
A0A0L7KPI6
+ More
S4NNF3 A0A212FEH9 A0A194RFK6 I4DKB4 A0A1J1I6C0 E2A1G9 A0A151K128 A0A0J7L3P3 A0A151J3L6 F4WHK2 A0A158N905 A0A026VSX7 A0A164PAT2 E2C438 A0A151IBK7 M7B729 A0A0P5FJH6 A0A0K8TS45 C1BSY2 A0A088AGW5 A0A2A3ETN3 V9IIP1 E9H6W9 A0A0P6C8H6 A0A0L7QWL4 A0A210Q0N7 V4A887 A0A0P6I4J1 B1H3F2 T1IHB3 C3Y0W8 F6V699 A0A1I8MQB1 A0A154P6M6 A0A3Q7PK43 C3YJY7 A0A1S3GB48 A0A3M7P239 G1MAQ8 A0A151MRJ1 A0A226MEY1 A0A2G9QIJ5 A0A286Y0R6 V5IAN6 L5LTW9 A0A1U7QYR3 A0A0P4VVP7 G1P2M1 A0A1U7R6N6 A0A1L8E839 D2I0T7 A0A2T7P0I9 M3YS00 A0A1L8EE83 A0A232EUR4 K7JA86 A0A250XVB3 A0A2P8XVC5 M3WGQ1 A0A1Y1N3A9 F6YLE9 F6W739 A0A2K6D0D3 A0A2K6JYG4 A0A2K5V9W8 G1RUR1 A0A2K5ZL84 A0A0D9S3H2 A0A2Y9EAC2 A0A2R9AYJ1 A0A096MRW1 A0A2K6RUA8 A0A2K5MA66 H0WUS6 A0A2K5IJL8 A0A2K6F995 H2N523 H2Q0F8 F6V169 B1AQP2 Q9UHV9 A0A034VYG9 L8YDV3 G5E1D4 L8HRH0 W5PID4 A0A212CGL8 A1A4P5 F1S192 Q862M6 G1SHF1 A0A226NUT8 I3M7R5 O70591
S4NNF3 A0A212FEH9 A0A194RFK6 I4DKB4 A0A1J1I6C0 E2A1G9 A0A151K128 A0A0J7L3P3 A0A151J3L6 F4WHK2 A0A158N905 A0A026VSX7 A0A164PAT2 E2C438 A0A151IBK7 M7B729 A0A0P5FJH6 A0A0K8TS45 C1BSY2 A0A088AGW5 A0A2A3ETN3 V9IIP1 E9H6W9 A0A0P6C8H6 A0A0L7QWL4 A0A210Q0N7 V4A887 A0A0P6I4J1 B1H3F2 T1IHB3 C3Y0W8 F6V699 A0A1I8MQB1 A0A154P6M6 A0A3Q7PK43 C3YJY7 A0A1S3GB48 A0A3M7P239 G1MAQ8 A0A151MRJ1 A0A226MEY1 A0A2G9QIJ5 A0A286Y0R6 V5IAN6 L5LTW9 A0A1U7QYR3 A0A0P4VVP7 G1P2M1 A0A1U7R6N6 A0A1L8E839 D2I0T7 A0A2T7P0I9 M3YS00 A0A1L8EE83 A0A232EUR4 K7JA86 A0A250XVB3 A0A2P8XVC5 M3WGQ1 A0A1Y1N3A9 F6YLE9 F6W739 A0A2K6D0D3 A0A2K6JYG4 A0A2K5V9W8 G1RUR1 A0A2K5ZL84 A0A0D9S3H2 A0A2Y9EAC2 A0A2R9AYJ1 A0A096MRW1 A0A2K6RUA8 A0A2K5MA66 H0WUS6 A0A2K5IJL8 A0A2K6F995 H2N523 H2Q0F8 F6V169 B1AQP2 Q9UHV9 A0A034VYG9 L8YDV3 G5E1D4 L8HRH0 W5PID4 A0A212CGL8 A1A4P5 F1S192 Q862M6 G1SHF1 A0A226NUT8 I3M7R5 O70591
Pubmed
19121390
28756777
26227816
23622113
22118469
26354079
+ More
22651552 20798317 21719571 21347285 24508170 30249741 23624526 26369729 21292972 28812685 23254933 18563158 20431018 25315136 30375419 20010809 22293439 21993624 28648823 20075255 28087693 29403074 17975172 28004739 17495919 19892987 22722832 25362486 16136131 17431167 22002653 25319552 11181995 10931946 11042152 15489334 9630229 17936702 21269460 25348373 23385571 22751099 20809919 30723633 12658628 24621616 21183079
22651552 20798317 21719571 21347285 24508170 30249741 23624526 26369729 21292972 28812685 23254933 18563158 20431018 25315136 30375419 20010809 22293439 21993624 28648823 20075255 28087693 29403074 17975172 28004739 17495919 19892987 22722832 25362486 16136131 17431167 22002653 25319552 11181995 10931946 11042152 15489334 9630229 17936702 21269460 25348373 23385571 22751099 20809919 30723633 12658628 24621616 21183079
EMBL
BABH01041181
ODYU01003125
SOQ41492.1
KZ150327
PZC71382.1
NWSH01003813
+ More
PCG65795.1 GDQN01006605 JAT84449.1 JTDY01007718 KOB64986.1 GAIX01012279 JAA80281.1 AGBW02008933 OWR52142.1 KQ460313 KPJ16080.1 AK401732 KQ459595 BAM18354.1 KPI95906.1 CVRI01000043 CRK95845.1 GL435766 EFN72642.1 KQ981229 KYN44341.1 LBMM01000785 KMQ97502.1 KQ980290 KYN16917.1 GL888161 EGI66356.1 ADTU01009011 KK108326 QOIP01000004 EZA46755.1 RLU23361.1 LRGB01002648 KZS06665.1 GL452364 EFN77424.1 KQ978095 KYM97023.1 KB565896 EMP27988.1 GDIQ01254465 JAJ97259.1 GDAI01000429 JAI17174.1 BT077711 BT120571 BT121940 HACA01009824 ACO12135.1 ADD24211.1 ADD38870.1 CDW27185.1 KZ288189 PBC34642.1 JR046359 AEY60179.1 GL732599 EFX72493.1 GDIP01007200 JAM96515.1 KQ414713 KOC62956.1 NEDP02005298 OWF42310.1 KB202591 ESO89496.1 GDIQ01011479 JAN83258.1 BC161373 AAI61373.1 JH429822 GG666479 EEN66101.1 AAMC01133308 KQ434827 KZC07596.1 GG666520 EEN59444.1 REGN01014039 RMZ93146.1 ACTA01073769 AKHW03005309 KYO27157.1 MCFN01001066 OXB53790.1 KV981459 PIO15335.1 AAKN02043132 AAKN02043133 GALX01000870 JAB67596.1 KB107990 ELK29487.1 GDRN01124704 JAI56694.1 AAPE02047223 AAPE02047224 AAPE02047225 AAPE02047226 AAPE02047227 AAPE02047228 AAPE02047229 GFDG01003975 JAV14824.1 GL193920 EFB27740.1 PZQS01000007 PVD26923.1 AEYP01088003 AEYP01088004 GFDG01001777 JAV17022.1 NNAY01002098 OXU22090.1 AAZX01006344 GFFV01004642 JAV35303.1 PYGN01001290 PSN35961.1 AANG04004454 GEZM01013807 JAV92319.1 AQIA01002038 ADFV01184071 ADFV01184072 ADFV01184073 ADFV01184074 ADFV01184075 AQIB01138704 AQIB01138705 AJFE02037540 AJFE02037541 AHZZ02017077 AAQR03039455 AAQR03039456 AAQR03039457 AAQR03039458 AAQR03039459 AAQR03039460 AAQR03039461 AAQR03039462 ABGA01295258 ABGA01295259 NDHI03003420 PNJ57076.1 AACZ04025384 GABF01007593 GABD01010716 NBAG03000493 JAA14552.1 JAA22384.1 PNI19817.1 JSUE03002659 JSUE03002660 JV043603 CM001253 AFI33674.1 EHH15435.1 AK315736 CH471121 BAG38091.1 EAW52658.1 AF117237 AF165883 AF151065 BC012464 BC047042 GAKP01012032 JAC46920.1 KB362198 ELV13279.1 JP287198 AEQ18144.1 JH883435 ELR46491.1 AMGL01005212 MKHE01000020 OWK05131.1 BC126786 AEMK02000022 DQIR01192919 HDB48396.1 AB098954 BAC56444.1 AAGW02000266 AWGT02000669 OXB71087.1 AGTP01116358 AGTP01116359 Y17393 BC026839 BC049606
PCG65795.1 GDQN01006605 JAT84449.1 JTDY01007718 KOB64986.1 GAIX01012279 JAA80281.1 AGBW02008933 OWR52142.1 KQ460313 KPJ16080.1 AK401732 KQ459595 BAM18354.1 KPI95906.1 CVRI01000043 CRK95845.1 GL435766 EFN72642.1 KQ981229 KYN44341.1 LBMM01000785 KMQ97502.1 KQ980290 KYN16917.1 GL888161 EGI66356.1 ADTU01009011 KK108326 QOIP01000004 EZA46755.1 RLU23361.1 LRGB01002648 KZS06665.1 GL452364 EFN77424.1 KQ978095 KYM97023.1 KB565896 EMP27988.1 GDIQ01254465 JAJ97259.1 GDAI01000429 JAI17174.1 BT077711 BT120571 BT121940 HACA01009824 ACO12135.1 ADD24211.1 ADD38870.1 CDW27185.1 KZ288189 PBC34642.1 JR046359 AEY60179.1 GL732599 EFX72493.1 GDIP01007200 JAM96515.1 KQ414713 KOC62956.1 NEDP02005298 OWF42310.1 KB202591 ESO89496.1 GDIQ01011479 JAN83258.1 BC161373 AAI61373.1 JH429822 GG666479 EEN66101.1 AAMC01133308 KQ434827 KZC07596.1 GG666520 EEN59444.1 REGN01014039 RMZ93146.1 ACTA01073769 AKHW03005309 KYO27157.1 MCFN01001066 OXB53790.1 KV981459 PIO15335.1 AAKN02043132 AAKN02043133 GALX01000870 JAB67596.1 KB107990 ELK29487.1 GDRN01124704 JAI56694.1 AAPE02047223 AAPE02047224 AAPE02047225 AAPE02047226 AAPE02047227 AAPE02047228 AAPE02047229 GFDG01003975 JAV14824.1 GL193920 EFB27740.1 PZQS01000007 PVD26923.1 AEYP01088003 AEYP01088004 GFDG01001777 JAV17022.1 NNAY01002098 OXU22090.1 AAZX01006344 GFFV01004642 JAV35303.1 PYGN01001290 PSN35961.1 AANG04004454 GEZM01013807 JAV92319.1 AQIA01002038 ADFV01184071 ADFV01184072 ADFV01184073 ADFV01184074 ADFV01184075 AQIB01138704 AQIB01138705 AJFE02037540 AJFE02037541 AHZZ02017077 AAQR03039455 AAQR03039456 AAQR03039457 AAQR03039458 AAQR03039459 AAQR03039460 AAQR03039461 AAQR03039462 ABGA01295258 ABGA01295259 NDHI03003420 PNJ57076.1 AACZ04025384 GABF01007593 GABD01010716 NBAG03000493 JAA14552.1 JAA22384.1 PNI19817.1 JSUE03002659 JSUE03002660 JV043603 CM001253 AFI33674.1 EHH15435.1 AK315736 CH471121 BAG38091.1 EAW52658.1 AF117237 AF165883 AF151065 BC012464 BC047042 GAKP01012032 JAC46920.1 KB362198 ELV13279.1 JP287198 AEQ18144.1 JH883435 ELR46491.1 AMGL01005212 MKHE01000020 OWK05131.1 BC126786 AEMK02000022 DQIR01192919 HDB48396.1 AB098954 BAC56444.1 AAGW02000266 AWGT02000669 OXB71087.1 AGTP01116358 AGTP01116359 Y17393 BC026839 BC049606
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000053268
+ More
UP000183832 UP000000311 UP000078541 UP000036403 UP000078492 UP000007755 UP000005205 UP000053097 UP000279307 UP000076858 UP000008237 UP000078542 UP000031443 UP000005203 UP000242457 UP000000305 UP000053825 UP000242188 UP000030746 UP000001554 UP000008143 UP000095301 UP000076502 UP000286641 UP000081671 UP000276133 UP000008912 UP000050525 UP000198323 UP000005447 UP000189706 UP000001074 UP000245119 UP000000715 UP000215335 UP000002358 UP000245037 UP000011712 UP000002280 UP000002281 UP000233120 UP000233180 UP000233100 UP000001073 UP000233140 UP000029965 UP000248480 UP000240080 UP000028761 UP000233200 UP000233060 UP000005225 UP000233080 UP000233160 UP000001595 UP000002277 UP000006718 UP000005640 UP000011518 UP000002356 UP000009136 UP000008227 UP000001811 UP000198419 UP000005215 UP000000589
UP000183832 UP000000311 UP000078541 UP000036403 UP000078492 UP000007755 UP000005205 UP000053097 UP000279307 UP000076858 UP000008237 UP000078542 UP000031443 UP000005203 UP000242457 UP000000305 UP000053825 UP000242188 UP000030746 UP000001554 UP000008143 UP000095301 UP000076502 UP000286641 UP000081671 UP000276133 UP000008912 UP000050525 UP000198323 UP000005447 UP000189706 UP000001074 UP000245119 UP000000715 UP000215335 UP000002358 UP000245037 UP000011712 UP000002280 UP000002281 UP000233120 UP000233180 UP000233100 UP000001073 UP000233140 UP000029965 UP000248480 UP000240080 UP000028761 UP000233200 UP000233060 UP000005225 UP000233080 UP000233160 UP000001595 UP000002277 UP000006718 UP000005640 UP000011518 UP000002356 UP000009136 UP000008227 UP000001811 UP000198419 UP000005215 UP000000589
Pfam
PF01920 Prefoldin_2
Gene 3D
ProteinModelPortal
H9JI41
A0A2H1VM24
A0A2W1B4U1
A0A2A4J137
A0A1E1WBX3
A0A0L7KPI6
+ More
S4NNF3 A0A212FEH9 A0A194RFK6 I4DKB4 A0A1J1I6C0 E2A1G9 A0A151K128 A0A0J7L3P3 A0A151J3L6 F4WHK2 A0A158N905 A0A026VSX7 A0A164PAT2 E2C438 A0A151IBK7 M7B729 A0A0P5FJH6 A0A0K8TS45 C1BSY2 A0A088AGW5 A0A2A3ETN3 V9IIP1 E9H6W9 A0A0P6C8H6 A0A0L7QWL4 A0A210Q0N7 V4A887 A0A0P6I4J1 B1H3F2 T1IHB3 C3Y0W8 F6V699 A0A1I8MQB1 A0A154P6M6 A0A3Q7PK43 C3YJY7 A0A1S3GB48 A0A3M7P239 G1MAQ8 A0A151MRJ1 A0A226MEY1 A0A2G9QIJ5 A0A286Y0R6 V5IAN6 L5LTW9 A0A1U7QYR3 A0A0P4VVP7 G1P2M1 A0A1U7R6N6 A0A1L8E839 D2I0T7 A0A2T7P0I9 M3YS00 A0A1L8EE83 A0A232EUR4 K7JA86 A0A250XVB3 A0A2P8XVC5 M3WGQ1 A0A1Y1N3A9 F6YLE9 F6W739 A0A2K6D0D3 A0A2K6JYG4 A0A2K5V9W8 G1RUR1 A0A2K5ZL84 A0A0D9S3H2 A0A2Y9EAC2 A0A2R9AYJ1 A0A096MRW1 A0A2K6RUA8 A0A2K5MA66 H0WUS6 A0A2K5IJL8 A0A2K6F995 H2N523 H2Q0F8 F6V169 B1AQP2 Q9UHV9 A0A034VYG9 L8YDV3 G5E1D4 L8HRH0 W5PID4 A0A212CGL8 A1A4P5 F1S192 Q862M6 G1SHF1 A0A226NUT8 I3M7R5 O70591
S4NNF3 A0A212FEH9 A0A194RFK6 I4DKB4 A0A1J1I6C0 E2A1G9 A0A151K128 A0A0J7L3P3 A0A151J3L6 F4WHK2 A0A158N905 A0A026VSX7 A0A164PAT2 E2C438 A0A151IBK7 M7B729 A0A0P5FJH6 A0A0K8TS45 C1BSY2 A0A088AGW5 A0A2A3ETN3 V9IIP1 E9H6W9 A0A0P6C8H6 A0A0L7QWL4 A0A210Q0N7 V4A887 A0A0P6I4J1 B1H3F2 T1IHB3 C3Y0W8 F6V699 A0A1I8MQB1 A0A154P6M6 A0A3Q7PK43 C3YJY7 A0A1S3GB48 A0A3M7P239 G1MAQ8 A0A151MRJ1 A0A226MEY1 A0A2G9QIJ5 A0A286Y0R6 V5IAN6 L5LTW9 A0A1U7QYR3 A0A0P4VVP7 G1P2M1 A0A1U7R6N6 A0A1L8E839 D2I0T7 A0A2T7P0I9 M3YS00 A0A1L8EE83 A0A232EUR4 K7JA86 A0A250XVB3 A0A2P8XVC5 M3WGQ1 A0A1Y1N3A9 F6YLE9 F6W739 A0A2K6D0D3 A0A2K6JYG4 A0A2K5V9W8 G1RUR1 A0A2K5ZL84 A0A0D9S3H2 A0A2Y9EAC2 A0A2R9AYJ1 A0A096MRW1 A0A2K6RUA8 A0A2K5MA66 H0WUS6 A0A2K5IJL8 A0A2K6F995 H2N523 H2Q0F8 F6V169 B1AQP2 Q9UHV9 A0A034VYG9 L8YDV3 G5E1D4 L8HRH0 W5PID4 A0A212CGL8 A1A4P5 F1S192 Q862M6 G1SHF1 A0A226NUT8 I3M7R5 O70591
Ontologies
GO
PANTHER
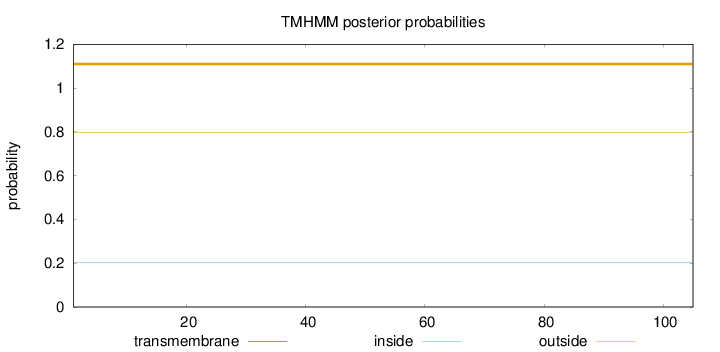
Topology
Subcellular location
Nucleus
Cytoplasm
Mitochondrion
Cytoplasm
Mitochondrion
Length:
105
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0013
Exp number, first 60 AAs:
0.0013
Total prob of N-in:
0.20193
outside
1 - 105
Population Genetic Test Statistics
Pi
231.034734
Theta
193.564008
Tajima's D
0.609255
CLR
0.089168
CSRT
0.546472676366182
Interpretation
Uncertain
