Gene
KWMTBOMO13480
Pre Gene Modal
BGIBMGA010874
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_cadherin-87A_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.44 Nuclear Reliability : 1.458
Sequence
CDS
ATGGCAGAGATCCTGCCGCGGCGAGGGGCGGACAGCTGGCCCGTACCCCTGCTGCTGCTTGCCGTGCTGCATCACGTCACGGCACAGCACGTGAACCGCGCGCCTCACTTCGTGCCGCAAGTCGGGGACATGTCTCACTTCACGCTCGCAGAGGACACGCCGGTCGGGACTCCCGTTTATCAACTCAAAGGCATCGATCCGGAGAACGATCAACTACGGTACTCGATATCTGGGCAACACTTGTCGGTGGACGCACTGAGTGGCGTGGTGACCCTCGTGCGGAGCCTCGACCGTGAGGAGCTGGACCAGCTCGAGGTCATCATCAGCATCACTGACGAGGCCAGGGAGTCATCGGAGCCGAACACCGTGTCTCTGAGGCGCGTGATCCCGGTCACCGACGTCAACGATAACCCGCCCGTCTTCCTCAATCGGCCCTACATAGTGAACATCAGCGAGGCGGCCCCGGTGGGGGCAGAGCTCCAGATAAATCCAAAAATACTAGTAACAGACCGAGACGCCGGACTAAACGCAAATCTCCTGGTGACGTGTTCCACCCGGGAGAAGGGCAGCGACGCTGAGGCGTGCGCCACCTTCAGGGTCGAGACTGTCGTGTTATCGTCGAGCGAGTACGAGGTGCGGCTGTTCGTGGACGCCGCGCTGGACTACGAGCGCAGGTCGGCGTACGTGGTGACGCTGCAGGCCGCCGACGCGTCCGAGTCCCCGCTGCGCGCCGCCGCCAGCGTCGCCGTCGCCGTGCGGGACGAGCAGGACCAGCCGCCCGTCTTCCTCAACGCGCCCTACTCCGCCACCGTGCCAGAGAACACGCCCACAAACACCAGTATACTAGAAATAATAGCAAAAGACGGTGACACAGCCAACCCGAGAGCAGTTCTTTTAACTATTGAGGGTGATACGGAAGGGTACTTCAAGTTGGTCCCCGAGAAGCCACTAGGGCGCGGTGTACTAGTCACTTCTGATAATCCAATCGACAGAGAAAGTGAAACTGTTTTACAAAACGGAGGAGTCTATACTTTCTTCATAAAGGCTACCGAACTAATAAACAACGAAGTGCCCTCGGACTACACGGTGACTTCGATCACCATCATAGTGACGGACGTCGATGATCACGCTCCGAGATTCAATAAAGATGTCTTCGATATTTCAATACCTGAAAACATAGAAAACGGCAGCCCCATACCAGGACTATCTATTTACGTTGAAGATGATGATATCGGACAAAACAGTAAATACGACTTGAAGTTGCGGGACGTGCTTAATTCGAGAGATGTATTCGCCGTGACTACGGAACACGGCGAAGGACGAACGCCAATCACCATCAAAGTAACAGATTCTTCCCGCTTAGACTACGACGTAGACAGTGATGAAGGCAGGTTGTTCAGTTTGGACATAGTGAGTTCACTGAATGACGTGGAGCTCTCGGCGGCTAGAGTCAATATTAAACTTTTAGATGTCAATGATAACGCTCCGGTTTTCGAACGGGCAACTTATAAATTCGATACTTTAGAAAATACTACCATTGGGTCAAAGGTAGGAGACGTTGCAGCGACAGACAAAGATTTCGGTATATTCGGAGAACTTGAGTATACTTTGACTGGCTTCGGATCCAATCTATTCAATACTGATAAGCAAAGAGGTGGATTGTATGTCCAGCAAGAGTTGGACTATGAGAAACAAAAGAGTTACAGTCTAACACTTTTCGCTAAAGATGGCGGAGGTAAAAGCTCATCCACTAGTATCTTCATCGATGTGTTAGATGTCAACGACAATGCTCCTATATTTGAGGCAACCGAGTACACCAGGACCATCAGAGACGGCGCCACCAGCTTCGAGCCACAACTCGTTCTGCGAGCTATAGACGTCGATGGGCCGACTCAAGGAAATGGTCGCATTAAATATTCAATAGAATCTGATAACAGCATTAACAATAAAGGCAGGGCGTTCGAAATAGATGAAGACACCGGAGAGCTCAGCATTGTGGATAAGTTAGATAGCATGGACACTCCGAGGGGACAATATGAACTTATTATTCGAGCAACTGATTACGGCATACCGGCCCTTCATAACGACACTCGAGTCTATATTAGAGTCGGAGTCCCGGGAAATCAACGGCCTACGTTCAAAGGGAACTATCATCTGTACAAACACACCGGGAACAATCAGACTCTAGATAATACTAATGAATACTCGTTTGATCTGAATCCTATGAACTACAAAGCAACTATTAAAGAAAATGCAAAGCCGGGTGACACCGTGACCAAGGTGACTGCCAGCGACCCGGACGGGCTCGACGCGCTGCTCGTGTACTCCATCGTGTCCGGCTCCAAAGACAACTTCGTTATTCATGAAAAAAGTGGACTGATAACCGTATCGAGCGACGCGAACCTGGACCGCGACGTGAGTCCGGAGCGCTACGAGCTCGTGGTGTCGGCCGTGGACAGCGGCACGCCCCTGCCGGAGACCGCCACCACCACGGTGCTCGTGCTCGTACAAGATGTTAACGATAAGCCGCCCAAGTTCAATTTCACGGAGTCCTCCACGTACATCTCTGAGAAGACCAAAGTCGGTGACGTTGTCGCTAAGATAGTAGCGTTCGACACTGATGTCAGTTCAAAACTTAAGTACAGTATTGTGAAACCGATGAGAGCGCTCTCTAAGGCTGGCGTCCAGTTACAGCCAAACTCACCCTATGATTACGCCAACTTGTTCCACATCGAGGAGGACACCGGGGAAATCCGGGTCAACGGAACTCTGGACTACAATCAAGCCAGCATAGTTATTCTAACCGTAAAGGTGATTGACGTCAACGCCGAAATAGATAAAGACAAACAGTTTGCCACCATCGATCACACGATCTACATACAGCCTTATGCTGATAAAAACCCCCAGTTTACGAATCCTGGTTGGAGTACGTCGCGGCCCTTGATTTATCACAAAATTAAAGAAGACCAACCTATCGGGAGCACCGTGCTCGTTCTCGCCGCTGAAGACCCAGTCTCGGGACATCTGGTGTCCAACTACAAAGTTATCAATTCAGAGACGGAATTACTTCAAGTGGATCCTCTAACGGGGCAAGTGGTTCTGACTAAGCACCTAGACTACGAGGAACTCACCAGCCCGAACCTAACACTGACCGTGAAGGCCACGAGTAGTGACGGCGCCCGGCACAGTGAAGCTAAGATAGTAATTGAAGTCATCAACATTAACGACAATCCACCGATATTCGAGAAAGAGGTTTACAGAGTGAGCGTTCTTGAATCTGTGAAGCATCCGGAACATATCGTGTCGGTGAAAGCAAATGACGCAGACGCCGTTCTTACTGAACAAGACAAACTTGAGGGATATTCGGATATACGGTATGCACTGCGCGGAGAGAATTCAGAGCTGTTCACCATCAACAACGTCACAGGAGCGATACAGATAGGCGCAGACAAGCATCTGGACCGGGAGCGGCAGTCGGTGCTGCGCGTGCTGGTGGAGGCGAGTGACGTGCCGCGCGGAGGCGCCCTCCGCCGCACGGCCGCCGCGCTGCTGCTCATAGACGTGCTCGACGTGGACGACAACGCGCCCGCCTTCGACAAGTCCCTCTACACCGCTGTAGTTCCCGAGAACGTGCCGGTGGGAATGGGCGTGATCAACGTGACGGCCACCGACCCTGACGAGGGGCTCGGGGGGGAGATCCGATACGAATTCCTTGACGAGGGGGAAGCTGCAGGCTTGTTTAGCATCGAGCCGCGGTCGGGGTGGGTTCACACAAGCGGCGCGCTGACCGGGCGAGGCCGCACGGCGCCGTACCGGCTGCTGGTGGGCGCGACGGACGCCGGCGGACACGCGGCTGACGCGTCGCTTGCCGTCTACATCGGTGACGTCAGCGCTAACGATGGCGTGCCGCGGTTCATCCGGCCCGCTCCTGGAGAACTACTCACCGTCAACGAGAACACAGCGCCGGGCACTGTGGTGTACGAGGTGGTGGCGACGGACCCAGACGACCCCACGCAGCCCTCCGGCCAGCTCCACTACTACATCCAGCAGAGCGATGACCTCACTCCCGCCTTCACCATCGAGAGGCGGACCGGTGTACTGCGCACGCGCGGCGCGCTGGACCGCGAGCGGCGCGCCGAGTACGCGCTGGTGGTGGTGGCCGAGGACGCGGGCCGCCCGCCGCTGCGAGCCACCGCGCTGCTGCGCCTCGCCGTCGCCGACCTCGACGACCACAAGCCGCACTTCGCCAGGAACCTCGACGACCCTCCAGTGGTGATGTTGGTGCAAGAGGAAGTCCCAATCGGTACCGAAATCGGGAGGCTGCGAGCCATTGATGAAGACATCGAAGAAAATGCAGCGATCGACTACGCCATCATATCGGGCAACGAGCTGGGCGCCGTGCGGGCGGAGCGCACCACCAGCAACGAGGCGCGGCTGGTGGCGGCCGCGCGGCTCGACCGGGAGCAGCTGGACCGCCTGCTCATCACCCTCAAATGTTTCAAGCACGGAACCACCCCGACACTCACCGACGCATACAACAGACTGGATCCGTCGCATATCCAGGTGTTGATAAAGGTGGAAGATTTGGACGACAACATACCCGAGTTCGTGGGCGGGAACGTGACGGTGGGCGTGCGGCTGAACGTGGCGCCGCACACCGCGGTGGTGACGCTGCGCGCCGCCGACGCCGACCCCGGGGCCGCCCCCCTCCGCTATCTCCTCGCCGCGGCCTCCTTTCAGTCACCTATTCAGGAGAAGTCGTTGTCCAATGTTAGTGATGTATTTTCACTGGACAACGTGACCGGAGAGCTGAAGATCGTGCAAAACCTCATTCACTACGCGGACGGGATATTCAGGCTGCGGGTCCTGGCTTGTAATTCGGACGAGGCCGATCGTTGCGGACAGGTGTGGGTCGAGGTGACAGTGGTTCGCGAGCGAGAGCTGCTGCGGCTGGAGGGCGCGGGCTGGTCGCGGCGGAGGCTGGGCGACCTGCAAGCTGCACTGGACGCAGTGCTGGCACCCCGAGCTCTTCGACTCCAACTGCATCTGCACGATGCCGCCGCACACACGCATCTCGGGCCGTGCTTCCAACTGCGCTCGGTGGAGACGGGCAGCGCCCAGACGGCGCGCGCGATGCTGGCGACGTTGCGCGCGCTGGACGCGGACGTGCGCCGCGTGCTGGAGCAGTACTCCGTGCGCAACGTGACGCGCTGCGGCCCGCCGCCCGCCCCGCCCCGCGCCGCGCACACCGCCCTCCTCGCCCTCGCCGCCGCGCTGCCCCTGGCCGCCCTCCTCACCGCCGTCACGCTGTGCTGTATGCACGATGCCGTGTCCTAA
Protein
MAEILPRRGADSWPVPLLLLAVLHHVTAQHVNRAPHFVPQVGDMSHFTLAEDTPVGTPVYQLKGIDPENDQLRYSISGQHLSVDALSGVVTLVRSLDREELDQLEVIISITDEARESSEPNTVSLRRVIPVTDVNDNPPVFLNRPYIVNISEAAPVGAELQINPKILVTDRDAGLNANLLVTCSTREKGSDAEACATFRVETVVLSSSEYEVRLFVDAALDYERRSAYVVTLQAADASESPLRAAASVAVAVRDEQDQPPVFLNAPYSATVPENTPTNTSILEIIAKDGDTANPRAVLLTIEGDTEGYFKLVPEKPLGRGVLVTSDNPIDRESETVLQNGGVYTFFIKATELINNEVPSDYTVTSITIIVTDVDDHAPRFNKDVFDISIPENIENGSPIPGLSIYVEDDDIGQNSKYDLKLRDVLNSRDVFAVTTEHGEGRTPITIKVTDSSRLDYDVDSDEGRLFSLDIVSSLNDVELSAARVNIKLLDVNDNAPVFERATYKFDTLENTTIGSKVGDVAATDKDFGIFGELEYTLTGFGSNLFNTDKQRGGLYVQQELDYEKQKSYSLTLFAKDGGGKSSSTSIFIDVLDVNDNAPIFEATEYTRTIRDGATSFEPQLVLRAIDVDGPTQGNGRIKYSIESDNSINNKGRAFEIDEDTGELSIVDKLDSMDTPRGQYELIIRATDYGIPALHNDTRVYIRVGVPGNQRPTFKGNYHLYKHTGNNQTLDNTNEYSFDLNPMNYKATIKENAKPGDTVTKVTASDPDGLDALLVYSIVSGSKDNFVIHEKSGLITVSSDANLDRDVSPERYELVVSAVDSGTPLPETATTTVLVLVQDVNDKPPKFNFTESSTYISEKTKVGDVVAKIVAFDTDVSSKLKYSIVKPMRALSKAGVQLQPNSPYDYANLFHIEEDTGEIRVNGTLDYNQASIVILTVKVIDVNAEIDKDKQFATIDHTIYIQPYADKNPQFTNPGWSTSRPLIYHKIKEDQPIGSTVLVLAAEDPVSGHLVSNYKVINSETELLQVDPLTGQVVLTKHLDYEELTSPNLTLTVKATSSDGARHSEAKIVIEVININDNPPIFEKEVYRVSVLESVKHPEHIVSVKANDADAVLTEQDKLEGYSDIRYALRGENSELFTINNVTGAIQIGADKHLDRERQSVLRVLVEASDVPRGGALRRTAAALLLIDVLDVDDNAPAFDKSLYTAVVPENVPVGMGVINVTATDPDEGLGGEIRYEFLDEGEAAGLFSIEPRSGWVHTSGALTGRGRTAPYRLLVGATDAGGHAADASLAVYIGDVSANDGVPRFIRPAPGELLTVNENTAPGTVVYEVVATDPDDPTQPSGQLHYYIQQSDDLTPAFTIERRTGVLRTRGALDRERRAEYALVVVAEDAGRPPLRATALLRLAVADLDDHKPHFARNLDDPPVVMLVQEEVPIGTEIGRLRAIDEDIEENAAIDYAIISGNELGAVRAERTTSNEARLVAAARLDREQLDRLLITLKCFKHGTTPTLTDAYNRLDPSHIQVLIKVEDLDDNIPEFVGGNVTVGVRLNVAPHTAVVTLRAADADPGAAPLRYLLAAASFQSPIQEKSLSNVSDVFSLDNVTGELKIVQNLIHYADGIFRLRVLACNSDEADRCGQVWVEVTVVRERELLRLEGAGWSRRRLGDLQAALDAVLAPRALRLQLHLHDAAAHTHLGPCFQLRSVETGSAQTARAMLATLRALDADVRRVLEQYSVRNVTRCGPPPAPPRAAHTALLALAAALPLAALLTAVTLCCMHDAVS
Summary
Uniprot
H9JMX1
A0A3S2NSB5
A0A2H1VMK1
A0A194PZC6
A0A194RE95
A0A0L7LFD3
+ More
A0A1S4FMU6 Q16VA8 B0WT78 A0A182YPW7 A0A182V666 Q7PPB2 A0A182LLG8 A0A182Q0Y0 A0A182MIZ1 A0A084VDI1 A0A3B0KN82 B3M6C0 A0A182WCI7 B4MMR2 A0A182RDG5 Q2LZH1 A0A182IQZ1 A0A1W4W4G9 A0A182F9R7 B4PJW5 W5JVV3 A0A026W912 B3NHW3 A0A182I143 A0A087ZPM8 B4LE98 A0A2A3E969 A0A182X8N1 E2CAA8 A0A0A1XSN6 A0A182UCG8 Q9VVG0 E1ZWE4 A0A195BS59 Q8IGX4 A0A0J9RX32 A0A158NUM4 F4WV67 A0A195EWL0 A0A195DRZ6 A0A034V178 A0A034V3B5 A0A154PKA4 A0A0M4EBR8 A0A1I8NNR5 A0A0K8WER0 A0A0K8VFS9 A0A195C950 W8B014 A0A0L0CGQ9 A0A067QQC1 E9J9Q4 A0A1I8MF45 A0A336M3U9 A0A182JW60 A0A232EY96 K7IRW7 A0A0C9QH22 A0A0M8ZS03 B4H6L4 A0A1B0FB48 A0A1B0AI13 A0A1A9VYS4 A0A1A9Y1I8 A0A0K8TY55 B4KZ25 A0A1A9WEA1 A0A2L1IQ97 A0A1B0BPW1 B4J244 A0A1B6CGW9 A0A3B0JZ06 A0A146LSS7 E0W375 T1IAK5 A0A2S2PYJ3 J9K5R4 A0A2H8U072 A0A1B0C4S7 A0A147BCT2 A0A2J7RE18 A0A0P5MDW1 A0A0P5GMS2 A0A1B6HJV7 A0A0N8C0C5 A0A0P5SB92 A0A0P5TYV8 E9HGV3 A0A165ADA7 A0A0P4Y2V4 A0A0P5N835 A0A0P5P3K3 A0A0P6G3M9
A0A1S4FMU6 Q16VA8 B0WT78 A0A182YPW7 A0A182V666 Q7PPB2 A0A182LLG8 A0A182Q0Y0 A0A182MIZ1 A0A084VDI1 A0A3B0KN82 B3M6C0 A0A182WCI7 B4MMR2 A0A182RDG5 Q2LZH1 A0A182IQZ1 A0A1W4W4G9 A0A182F9R7 B4PJW5 W5JVV3 A0A026W912 B3NHW3 A0A182I143 A0A087ZPM8 B4LE98 A0A2A3E969 A0A182X8N1 E2CAA8 A0A0A1XSN6 A0A182UCG8 Q9VVG0 E1ZWE4 A0A195BS59 Q8IGX4 A0A0J9RX32 A0A158NUM4 F4WV67 A0A195EWL0 A0A195DRZ6 A0A034V178 A0A034V3B5 A0A154PKA4 A0A0M4EBR8 A0A1I8NNR5 A0A0K8WER0 A0A0K8VFS9 A0A195C950 W8B014 A0A0L0CGQ9 A0A067QQC1 E9J9Q4 A0A1I8MF45 A0A336M3U9 A0A182JW60 A0A232EY96 K7IRW7 A0A0C9QH22 A0A0M8ZS03 B4H6L4 A0A1B0FB48 A0A1B0AI13 A0A1A9VYS4 A0A1A9Y1I8 A0A0K8TY55 B4KZ25 A0A1A9WEA1 A0A2L1IQ97 A0A1B0BPW1 B4J244 A0A1B6CGW9 A0A3B0JZ06 A0A146LSS7 E0W375 T1IAK5 A0A2S2PYJ3 J9K5R4 A0A2H8U072 A0A1B0C4S7 A0A147BCT2 A0A2J7RE18 A0A0P5MDW1 A0A0P5GMS2 A0A1B6HJV7 A0A0N8C0C5 A0A0P5SB92 A0A0P5TYV8 E9HGV3 A0A165ADA7 A0A0P4Y2V4 A0A0P5N835 A0A0P5P3K3 A0A0P6G3M9
Pubmed
19121390
26354079
26227816
17510324
25244985
12364791
+ More
14747013 17210077 20966253 24438588 17994087 15632085 23185243 17550304 20920257 23761445 24508170 30249741 18057021 20798317 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 21347285 21719571 25348373 24495485 26108605 24845553 21282665 25315136 28648823 20075255 29415259 26823975 20566863 29652888 21292972
14747013 17210077 20966253 24438588 17994087 15632085 23185243 17550304 20920257 23761445 24508170 30249741 18057021 20798317 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 21347285 21719571 25348373 24495485 26108605 24845553 21282665 25315136 28648823 20075255 29415259 26823975 20566863 29652888 21292972
EMBL
BABH01028984
BABH01028985
BABH01028986
RSAL01000106
RVE47300.1
ODYU01003376
+ More
SOQ42059.1 KQ459586 KPI98094.1 KQ460323 KPJ15912.1 JTDY01001305 KOB74248.1 CH477597 EAT38525.1 DS232081 EDS34227.1 AAAB01008960 EAA10759.5 AXCN02000346 AXCM01007992 ATLV01011338 KE524664 KFB36025.1 OUUW01000012 SPP87306.1 CH902618 EDV39740.1 CH963847 EDW73468.1 CH379069 EAL29536.2 KRT08111.1 KRT08112.1 KRT08113.1 CM000159 EDW94734.1 KRK02021.1 KRK02022.1 KRK02023.1 ADMH02000262 ETN67210.1 KK107324 QOIP01000006 EZA52550.1 RLU22000.1 CH954178 EDV51978.1 KQS44050.1 KQS44051.1 KQS44052.1 APCN01000030 CH940647 EDW69054.1 KRF84151.1 KRF84152.1 KRF84153.1 KRF84154.1 KRF84155.1 KZ288341 PBC27729.1 GL453958 EFN75120.1 GBXI01000402 JAD13890.1 AE014296 BT150474 AAF49351.1 AAN11706.1 AFH04485.1 AKG26251.1 GL434819 EFN74490.1 KQ976417 KYM89867.1 BT001543 AAN71298.1 CM002912 KMZ00217.1 ADTU01026618 ADTU01026619 ADTU01026620 GL888384 EGI61955.1 KQ981953 KYN32536.1 KQ980530 KYN15621.1 GAKP01021926 JAC37026.1 GAKP01021928 JAC37024.1 KQ434943 KZC12275.1 CP012525 ALC44966.1 GDHF01002785 JAI49529.1 GDHF01014909 JAI37405.1 KQ978164 KYM96638.1 GAMC01014688 JAB91867.1 JRES01000428 KNC31372.1 KK853136 KDR10809.1 GL769399 EFZ10481.1 UFQT01000522 SSX24925.1 NNAY01001659 OXU23295.1 GBYB01013943 JAG83710.1 KQ435878 KOX69950.1 CH479214 EDW33442.1 CCAG010007618 GDHF01032910 JAI19404.1 CH933809 EDW17822.2 MF741664 AVD96947.1 JXJN01018247 CH916366 EDV96995.1 GEDC01024649 JAS12649.1 SPP87307.1 GDHC01007785 JAQ10844.1 DS235882 EEB20081.1 ACPB03005336 GGMS01001384 MBY70587.1 ABLF02034795 GFXV01008109 MBW19914.1 JXJN01025643 GEGO01007132 JAR88272.1 NEVH01005279 PNF39072.1 GDIQ01190587 GDIQ01168794 GDIQ01144356 GDIQ01121111 GDIQ01121110 JAK82931.1 GDIQ01239471 JAK12254.1 GECU01032747 JAS74959.1 GDIQ01127259 JAL24467.1 GDIP01142521 JAL61193.1 GDIP01124572 JAL79142.1 GL732643 EFX69052.1 LRGB01000626 KZS17483.1 GDIP01235557 JAI87844.1 GDIQ01149701 JAL02025.1 GDIQ01138510 JAL13216.1 GDIQ01041294 JAN53443.1
SOQ42059.1 KQ459586 KPI98094.1 KQ460323 KPJ15912.1 JTDY01001305 KOB74248.1 CH477597 EAT38525.1 DS232081 EDS34227.1 AAAB01008960 EAA10759.5 AXCN02000346 AXCM01007992 ATLV01011338 KE524664 KFB36025.1 OUUW01000012 SPP87306.1 CH902618 EDV39740.1 CH963847 EDW73468.1 CH379069 EAL29536.2 KRT08111.1 KRT08112.1 KRT08113.1 CM000159 EDW94734.1 KRK02021.1 KRK02022.1 KRK02023.1 ADMH02000262 ETN67210.1 KK107324 QOIP01000006 EZA52550.1 RLU22000.1 CH954178 EDV51978.1 KQS44050.1 KQS44051.1 KQS44052.1 APCN01000030 CH940647 EDW69054.1 KRF84151.1 KRF84152.1 KRF84153.1 KRF84154.1 KRF84155.1 KZ288341 PBC27729.1 GL453958 EFN75120.1 GBXI01000402 JAD13890.1 AE014296 BT150474 AAF49351.1 AAN11706.1 AFH04485.1 AKG26251.1 GL434819 EFN74490.1 KQ976417 KYM89867.1 BT001543 AAN71298.1 CM002912 KMZ00217.1 ADTU01026618 ADTU01026619 ADTU01026620 GL888384 EGI61955.1 KQ981953 KYN32536.1 KQ980530 KYN15621.1 GAKP01021926 JAC37026.1 GAKP01021928 JAC37024.1 KQ434943 KZC12275.1 CP012525 ALC44966.1 GDHF01002785 JAI49529.1 GDHF01014909 JAI37405.1 KQ978164 KYM96638.1 GAMC01014688 JAB91867.1 JRES01000428 KNC31372.1 KK853136 KDR10809.1 GL769399 EFZ10481.1 UFQT01000522 SSX24925.1 NNAY01001659 OXU23295.1 GBYB01013943 JAG83710.1 KQ435878 KOX69950.1 CH479214 EDW33442.1 CCAG010007618 GDHF01032910 JAI19404.1 CH933809 EDW17822.2 MF741664 AVD96947.1 JXJN01018247 CH916366 EDV96995.1 GEDC01024649 JAS12649.1 SPP87307.1 GDHC01007785 JAQ10844.1 DS235882 EEB20081.1 ACPB03005336 GGMS01001384 MBY70587.1 ABLF02034795 GFXV01008109 MBW19914.1 JXJN01025643 GEGO01007132 JAR88272.1 NEVH01005279 PNF39072.1 GDIQ01190587 GDIQ01168794 GDIQ01144356 GDIQ01121111 GDIQ01121110 JAK82931.1 GDIQ01239471 JAK12254.1 GECU01032747 JAS74959.1 GDIQ01127259 JAL24467.1 GDIP01142521 JAL61193.1 GDIP01124572 JAL79142.1 GL732643 EFX69052.1 LRGB01000626 KZS17483.1 GDIP01235557 JAI87844.1 GDIQ01149701 JAL02025.1 GDIQ01138510 JAL13216.1 GDIQ01041294 JAN53443.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000037510
UP000008820
+ More
UP000002320 UP000076408 UP000075903 UP000007062 UP000075882 UP000075886 UP000075883 UP000030765 UP000268350 UP000007801 UP000075920 UP000007798 UP000075900 UP000001819 UP000075880 UP000192221 UP000069272 UP000002282 UP000000673 UP000053097 UP000279307 UP000008711 UP000075840 UP000005203 UP000008792 UP000242457 UP000076407 UP000008237 UP000075902 UP000000803 UP000000311 UP000078540 UP000005205 UP000007755 UP000078541 UP000078492 UP000076502 UP000092553 UP000095300 UP000078542 UP000037069 UP000027135 UP000095301 UP000075881 UP000215335 UP000002358 UP000053105 UP000008744 UP000092444 UP000092445 UP000078200 UP000092443 UP000009192 UP000091820 UP000092460 UP000001070 UP000009046 UP000015103 UP000007819 UP000235965 UP000000305 UP000076858
UP000002320 UP000076408 UP000075903 UP000007062 UP000075882 UP000075886 UP000075883 UP000030765 UP000268350 UP000007801 UP000075920 UP000007798 UP000075900 UP000001819 UP000075880 UP000192221 UP000069272 UP000002282 UP000000673 UP000053097 UP000279307 UP000008711 UP000075840 UP000005203 UP000008792 UP000242457 UP000076407 UP000008237 UP000075902 UP000000803 UP000000311 UP000078540 UP000005205 UP000007755 UP000078541 UP000078492 UP000076502 UP000092553 UP000095300 UP000078542 UP000037069 UP000027135 UP000095301 UP000075881 UP000215335 UP000002358 UP000053105 UP000008744 UP000092444 UP000092445 UP000078200 UP000092443 UP000009192 UP000091820 UP000092460 UP000001070 UP000009046 UP000015103 UP000007819 UP000235965 UP000000305 UP000076858
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JMX1
A0A3S2NSB5
A0A2H1VMK1
A0A194PZC6
A0A194RE95
A0A0L7LFD3
+ More
A0A1S4FMU6 Q16VA8 B0WT78 A0A182YPW7 A0A182V666 Q7PPB2 A0A182LLG8 A0A182Q0Y0 A0A182MIZ1 A0A084VDI1 A0A3B0KN82 B3M6C0 A0A182WCI7 B4MMR2 A0A182RDG5 Q2LZH1 A0A182IQZ1 A0A1W4W4G9 A0A182F9R7 B4PJW5 W5JVV3 A0A026W912 B3NHW3 A0A182I143 A0A087ZPM8 B4LE98 A0A2A3E969 A0A182X8N1 E2CAA8 A0A0A1XSN6 A0A182UCG8 Q9VVG0 E1ZWE4 A0A195BS59 Q8IGX4 A0A0J9RX32 A0A158NUM4 F4WV67 A0A195EWL0 A0A195DRZ6 A0A034V178 A0A034V3B5 A0A154PKA4 A0A0M4EBR8 A0A1I8NNR5 A0A0K8WER0 A0A0K8VFS9 A0A195C950 W8B014 A0A0L0CGQ9 A0A067QQC1 E9J9Q4 A0A1I8MF45 A0A336M3U9 A0A182JW60 A0A232EY96 K7IRW7 A0A0C9QH22 A0A0M8ZS03 B4H6L4 A0A1B0FB48 A0A1B0AI13 A0A1A9VYS4 A0A1A9Y1I8 A0A0K8TY55 B4KZ25 A0A1A9WEA1 A0A2L1IQ97 A0A1B0BPW1 B4J244 A0A1B6CGW9 A0A3B0JZ06 A0A146LSS7 E0W375 T1IAK5 A0A2S2PYJ3 J9K5R4 A0A2H8U072 A0A1B0C4S7 A0A147BCT2 A0A2J7RE18 A0A0P5MDW1 A0A0P5GMS2 A0A1B6HJV7 A0A0N8C0C5 A0A0P5SB92 A0A0P5TYV8 E9HGV3 A0A165ADA7 A0A0P4Y2V4 A0A0P5N835 A0A0P5P3K3 A0A0P6G3M9
A0A1S4FMU6 Q16VA8 B0WT78 A0A182YPW7 A0A182V666 Q7PPB2 A0A182LLG8 A0A182Q0Y0 A0A182MIZ1 A0A084VDI1 A0A3B0KN82 B3M6C0 A0A182WCI7 B4MMR2 A0A182RDG5 Q2LZH1 A0A182IQZ1 A0A1W4W4G9 A0A182F9R7 B4PJW5 W5JVV3 A0A026W912 B3NHW3 A0A182I143 A0A087ZPM8 B4LE98 A0A2A3E969 A0A182X8N1 E2CAA8 A0A0A1XSN6 A0A182UCG8 Q9VVG0 E1ZWE4 A0A195BS59 Q8IGX4 A0A0J9RX32 A0A158NUM4 F4WV67 A0A195EWL0 A0A195DRZ6 A0A034V178 A0A034V3B5 A0A154PKA4 A0A0M4EBR8 A0A1I8NNR5 A0A0K8WER0 A0A0K8VFS9 A0A195C950 W8B014 A0A0L0CGQ9 A0A067QQC1 E9J9Q4 A0A1I8MF45 A0A336M3U9 A0A182JW60 A0A232EY96 K7IRW7 A0A0C9QH22 A0A0M8ZS03 B4H6L4 A0A1B0FB48 A0A1B0AI13 A0A1A9VYS4 A0A1A9Y1I8 A0A0K8TY55 B4KZ25 A0A1A9WEA1 A0A2L1IQ97 A0A1B0BPW1 B4J244 A0A1B6CGW9 A0A3B0JZ06 A0A146LSS7 E0W375 T1IAK5 A0A2S2PYJ3 J9K5R4 A0A2H8U072 A0A1B0C4S7 A0A147BCT2 A0A2J7RE18 A0A0P5MDW1 A0A0P5GMS2 A0A1B6HJV7 A0A0N8C0C5 A0A0P5SB92 A0A0P5TYV8 E9HGV3 A0A165ADA7 A0A0P4Y2V4 A0A0P5N835 A0A0P5P3K3 A0A0P6G3M9
PDB
6E6B
E-value=1.0379e-29,
Score=330
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 28,
Likelihood: 0.961846
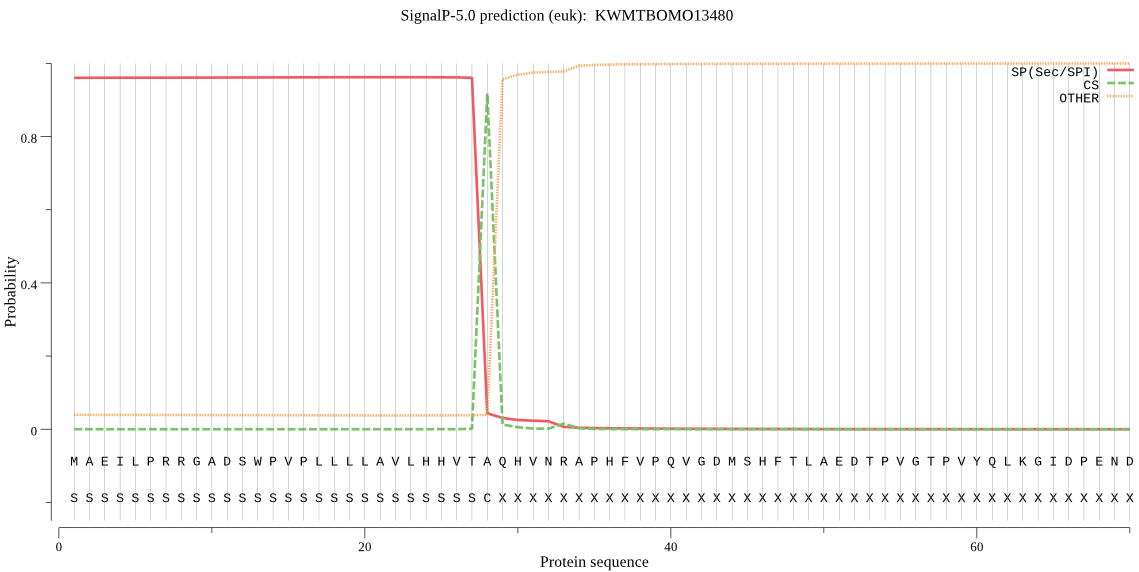
Length:
1778
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.75971
Exp number, first 60 AAs:
0.11303
Total prob of N-in:
0.00603
outside
1 - 1749
TMhelix
1750 - 1772
inside
1773 - 1778

Population Genetic Test Statistics
Pi
25.206408
Theta
20.485139
Tajima's D
-1.23017
CLR
0.428741
CSRT
0.101194940252987
Interpretation
Uncertain
