Gene
KWMTBOMO13457
Pre Gene Modal
BGIBMGA010885
Annotation
PREDICTED:_radial_spoke_head_protein_3_homolog_B_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.386
Sequence
CDS
ATGAAAGGCATTACCTTAATACCTCTCTTCGATTTTGACTTGGAAGTGCAGCCGATAATTGAAGTGCTGATCGGCAAAACGACAGAGCAGGCCTTGGCTGAAGTGGCCCAGGAGGAGGAGCTGGCGACACTGCGCGAGCAGCAGCGCCGGTACTGCGAGCTGCGGGACGCCGAGCGCGCCGAGAGGCAGAGGCTCGCCGCCCAACACCTGAGGCTGCACGAAGAGAAGGAAAGACGGGTCACCGAGGCGGAAGCCGCCCAAATAGCGGCCATTGAAGCGAGAGAGCGTACCGCCGCTGCTGTACTCGTGCAAGGATACATAGCTGAACTCCTGCCTTCAGTGCTGGCTGGTTTGAGAGACCAAGGATATCTGGTGGAGAGAATAGAGAGAGGAGTCGATGAAGAGTTCATGCCTTGGCTGGTGAAGGAAGTGTCACTGGAGATCGAGTCCATCATCACGAGTCGTGACGTCATCACGGAAATCGTACGTGACGTGGTGGAAGCCAGAGGAGAACTGTACAGAAACGTTGCTGAGAGAGAGGAGAATGAACAGGGGAGGAACGAGCCCGAGGACAGCATCACTCACCCCTCCGCCGTGTCTGAGGAGGGGGAGGAAGAGCGTGAGTATACTTGTTTATAA
Protein
MKGITLIPLFDFDLEVQPIIEVLIGKTTEQALAEVAQEEELATLREQQRRYCELRDAERAERQRLAAQHLRLHEEKERRVTEAEAAQIAAIEARERTAAAVLVQGYIAELLPSVLAGLRDQGYLVERIERGVDEEFMPWLVKEVSLEIESIITSRDVITEIVRDVVEARGELYRNVAEREENEQGRNEPEDSITHPSAVSEEGEEEREYTCL
Summary
Uniprot
H9JMY2
A0A2H1W6G9
A0A2W1BML9
A0A212F3B6
A0A194PXM7
A0A3S2NMJ8
+ More
D6WLL4 A0A1Y1NE13 N6UCS1 A0A0T6BFX0 E2BMZ0 A0A0L7RIW4 E0W3K1 A0A195CJU2 K7J0L8 A0A232ERE5 A0A1B6GIY0 A0A1B6DKY5 A0A1B6GCM0 A0A088AHV1 A0A0J7KWG0 A0A026WL04 F4WTL0 A0A3L8D6W2 A0A0A9XPR7 A0A158NGF8 A0A1B6MSV4 A0A0A9XLI6 A0A0K8SRW7 A0A195F1X6 A0A154PS47 E2A9J4 A0A151XBA2 A0A195E7A3 E9I939 A0A0C9QQB3 A0A0C9RC71 A0A0C9PWL1 A0A224XRV0 A0A151I145 A0A2I2Y252 Q8T898 H2XJZ5 A0A0N0BIU4 A0A3B4TER1 T1HAT2 A0A3B4YVG8 G3QMQ1 A0A267DEC2 A0A3B4ZH51 A0A1I8HW35 A0A2G8LDS9 A0A3Q1K2A6 A0A2I3NBN1 A0A2A3EA67 A0A2K5MR19 A0A2I3N1X3 A0A3Q3RR07 A0A2K6MRV6 A0A337SAG0 A0A0F8C8K1 I3JFD2 A0A2K6P9B0 A0A2K5ZYV5 A0A096N0X5 A0A2F0BJ09 A0A3Q1BH65 A0A091DRE1 B0CM43 A0A3P8S2Q7 M3ZBJ3 A0A091I0V5 A0A3B4H8N2 A0A3Q2VUW3 A0A3B4HB00 A0A3P9AUF2 B3RY22 A0A3P8R0P4 A0A369SLD1 A0A2K6D0C1 A0A2K5VVN2 F7GJB5 A0A2K6P9B7 A0A2I3M6Y6 G5BCM0 A0A3P8X401 A0A2Y9FBC9 A0A1U7SRI2 A0A3M6T605 Q4R807 A0A2R8ZKF1 A0A2I3T3D0 A0A0C4DG29 A0A2J8XAZ6 A0A2K5MR14 Q95KH8 A0A1D5Q281 A0A3Q4I1F9
D6WLL4 A0A1Y1NE13 N6UCS1 A0A0T6BFX0 E2BMZ0 A0A0L7RIW4 E0W3K1 A0A195CJU2 K7J0L8 A0A232ERE5 A0A1B6GIY0 A0A1B6DKY5 A0A1B6GCM0 A0A088AHV1 A0A0J7KWG0 A0A026WL04 F4WTL0 A0A3L8D6W2 A0A0A9XPR7 A0A158NGF8 A0A1B6MSV4 A0A0A9XLI6 A0A0K8SRW7 A0A195F1X6 A0A154PS47 E2A9J4 A0A151XBA2 A0A195E7A3 E9I939 A0A0C9QQB3 A0A0C9RC71 A0A0C9PWL1 A0A224XRV0 A0A151I145 A0A2I2Y252 Q8T898 H2XJZ5 A0A0N0BIU4 A0A3B4TER1 T1HAT2 A0A3B4YVG8 G3QMQ1 A0A267DEC2 A0A3B4ZH51 A0A1I8HW35 A0A2G8LDS9 A0A3Q1K2A6 A0A2I3NBN1 A0A2A3EA67 A0A2K5MR19 A0A2I3N1X3 A0A3Q3RR07 A0A2K6MRV6 A0A337SAG0 A0A0F8C8K1 I3JFD2 A0A2K6P9B0 A0A2K5ZYV5 A0A096N0X5 A0A2F0BJ09 A0A3Q1BH65 A0A091DRE1 B0CM43 A0A3P8S2Q7 M3ZBJ3 A0A091I0V5 A0A3B4H8N2 A0A3Q2VUW3 A0A3B4HB00 A0A3P9AUF2 B3RY22 A0A3P8R0P4 A0A369SLD1 A0A2K6D0C1 A0A2K5VVN2 F7GJB5 A0A2K6P9B7 A0A2I3M6Y6 G5BCM0 A0A3P8X401 A0A2Y9FBC9 A0A1U7SRI2 A0A3M6T605 Q4R807 A0A2R8ZKF1 A0A2I3T3D0 A0A0C4DG29 A0A2J8XAZ6 A0A2K5MR14 Q95KH8 A0A1D5Q281 A0A3Q4I1F9
Pubmed
19121390
28756777
22118469
26354079
18362917
19820115
+ More
28004739 23537049 20798317 20566863 20075255 28648823 24508170 21719571 30249741 25401762 21347285 26823975 21282665 22398555 12589069 12481130 15114417 26392545 29023486 17975172 25835551 25186727 25362486 18719581 30042472 17431167 21993625 24487278 30382153 15944441 22722832 16136131 14574404
28004739 23537049 20798317 20566863 20075255 28648823 24508170 21719571 30249741 25401762 21347285 26823975 21282665 22398555 12589069 12481130 15114417 26392545 29023486 17975172 25835551 25186727 25362486 18719581 30042472 17431167 21993625 24487278 30382153 15944441 22722832 16136131 14574404
EMBL
BABH01029052
BABH01029053
BABH01029054
ODYU01006581
SOQ48546.1
KZ149966
+ More
PZC76178.1 AGBW02010583 OWR48230.1 KQ459586 KPI98077.1 RSAL01008030 RVE39812.1 KQ971343 EFA04128.2 GEZM01005363 JAV96204.1 APGK01034095 KB740904 KB632006 ENN78431.1 ERL87976.1 LJIG01000723 KRT86217.1 GL449382 EFN82894.1 KQ414583 KOC70748.1 DS235882 EEB20207.1 KQ977721 KYN00339.1 NNAY01002619 OXU20928.1 GECZ01007373 JAS62396.1 GEDC01010986 JAS26312.1 GECZ01009588 JAS60181.1 LBMM01002579 KMQ94636.1 KK107185 EZA55789.1 GL888341 EGI62431.1 QOIP01000012 RLU16004.1 GBHO01022771 GBRD01009730 JAG20833.1 JAG56094.1 ADTU01014938 GEBQ01000994 JAT38983.1 GBHO01022770 JAG20834.1 GBRD01009732 GDHC01006331 JAG56092.1 JAQ12298.1 KQ981864 KYN34172.1 KQ435119 KZC14729.1 GL437917 EFN69884.1 KQ982335 KYQ57627.1 KQ979568 KYN20976.1 GL761674 EFZ22918.1 GBYB01005889 JAG75656.1 GBYB01004375 GBYB01005890 JAG74142.1 JAG75657.1 GBYB01005888 JAG75655.1 GFTR01005717 JAW10709.1 KQ976580 KYM79922.1 CABD030048713 CABD030048714 CABD030048715 CABD030048716 AB074930 BAB85844.1 EAAA01001637 KQ435726 KOX78162.1 ACPB03015547 ACPB03015548 ACPB03015549 NIVC01004599 PAA47047.1 NIVC01002314 NIVC01000446 PAA58963.1 PAA82813.1 MRZV01000113 PIK58401.1 AHZZ02026000 AHZZ02026001 KZ288310 PBC28608.1 AANG04000169 KQ042552 KKF13810.1 AERX01015266 NTJE010229635 MBW02979.1 KN122124 KFO32835.1 DP000548 ABY56292.1 ADFV01067976 ADFV01067977 ADFV01067978 ADFV01067979 KL218012 KFP01053.1 DS985245 EDV24527.1 NOWV01000006 RDD46833.1 AQIA01049359 JSUE03032532 JH169546 EHB07031.1 RCHS01004216 RMX36801.1 AB168654 BAE00765.1 AJFE02021400 AJFE02021401 AACZ04028794 NBAG03000358 PNI35439.1 AL035530 NDHI03003369 PNJ79172.1 AB060862 BAB46879.1
PZC76178.1 AGBW02010583 OWR48230.1 KQ459586 KPI98077.1 RSAL01008030 RVE39812.1 KQ971343 EFA04128.2 GEZM01005363 JAV96204.1 APGK01034095 KB740904 KB632006 ENN78431.1 ERL87976.1 LJIG01000723 KRT86217.1 GL449382 EFN82894.1 KQ414583 KOC70748.1 DS235882 EEB20207.1 KQ977721 KYN00339.1 NNAY01002619 OXU20928.1 GECZ01007373 JAS62396.1 GEDC01010986 JAS26312.1 GECZ01009588 JAS60181.1 LBMM01002579 KMQ94636.1 KK107185 EZA55789.1 GL888341 EGI62431.1 QOIP01000012 RLU16004.1 GBHO01022771 GBRD01009730 JAG20833.1 JAG56094.1 ADTU01014938 GEBQ01000994 JAT38983.1 GBHO01022770 JAG20834.1 GBRD01009732 GDHC01006331 JAG56092.1 JAQ12298.1 KQ981864 KYN34172.1 KQ435119 KZC14729.1 GL437917 EFN69884.1 KQ982335 KYQ57627.1 KQ979568 KYN20976.1 GL761674 EFZ22918.1 GBYB01005889 JAG75656.1 GBYB01004375 GBYB01005890 JAG74142.1 JAG75657.1 GBYB01005888 JAG75655.1 GFTR01005717 JAW10709.1 KQ976580 KYM79922.1 CABD030048713 CABD030048714 CABD030048715 CABD030048716 AB074930 BAB85844.1 EAAA01001637 KQ435726 KOX78162.1 ACPB03015547 ACPB03015548 ACPB03015549 NIVC01004599 PAA47047.1 NIVC01002314 NIVC01000446 PAA58963.1 PAA82813.1 MRZV01000113 PIK58401.1 AHZZ02026000 AHZZ02026001 KZ288310 PBC28608.1 AANG04000169 KQ042552 KKF13810.1 AERX01015266 NTJE010229635 MBW02979.1 KN122124 KFO32835.1 DP000548 ABY56292.1 ADFV01067976 ADFV01067977 ADFV01067978 ADFV01067979 KL218012 KFP01053.1 DS985245 EDV24527.1 NOWV01000006 RDD46833.1 AQIA01049359 JSUE03032532 JH169546 EHB07031.1 RCHS01004216 RMX36801.1 AB168654 BAE00765.1 AJFE02021400 AJFE02021401 AACZ04028794 NBAG03000358 PNI35439.1 AL035530 NDHI03003369 PNJ79172.1 AB060862 BAB46879.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000283053
UP000007266
UP000019118
+ More
UP000030742 UP000008237 UP000053825 UP000009046 UP000078542 UP000002358 UP000215335 UP000005203 UP000036403 UP000053097 UP000007755 UP000279307 UP000005205 UP000078541 UP000076502 UP000000311 UP000075809 UP000078492 UP000078540 UP000001519 UP000008144 UP000053105 UP000261420 UP000015103 UP000261360 UP000215902 UP000261400 UP000095280 UP000230750 UP000265040 UP000028761 UP000242457 UP000233060 UP000261640 UP000233180 UP000011712 UP000005207 UP000233200 UP000233140 UP000257160 UP000028990 UP000265080 UP000001073 UP000054308 UP000261460 UP000264840 UP000265160 UP000009022 UP000265100 UP000253843 UP000233120 UP000233100 UP000006718 UP000006813 UP000265120 UP000189704 UP000275408 UP000240080 UP000002277 UP000005640 UP000261580
UP000030742 UP000008237 UP000053825 UP000009046 UP000078542 UP000002358 UP000215335 UP000005203 UP000036403 UP000053097 UP000007755 UP000279307 UP000005205 UP000078541 UP000076502 UP000000311 UP000075809 UP000078492 UP000078540 UP000001519 UP000008144 UP000053105 UP000261420 UP000015103 UP000261360 UP000215902 UP000261400 UP000095280 UP000230750 UP000265040 UP000028761 UP000242457 UP000233060 UP000261640 UP000233180 UP000011712 UP000005207 UP000233200 UP000233140 UP000257160 UP000028990 UP000265080 UP000001073 UP000054308 UP000261460 UP000264840 UP000265160 UP000009022 UP000265100 UP000253843 UP000233120 UP000233100 UP000006718 UP000006813 UP000265120 UP000189704 UP000275408 UP000240080 UP000002277 UP000005640 UP000261580
PRIDE
Interpro
IPR009290
Radial_spoke_3
+ More
IPR041789 ERM_FERM_C
IPR018979 FERM_N
IPR019747 FERM_CS
IPR019748 FERM_central
IPR000798 Ez/rad/moesin-like
IPR011993 PH-like_dom_sf
IPR018980 FERM_PH-like_C
IPR000299 FERM_domain
IPR029071 Ubiquitin-like_domsf
IPR008954 Moesin_tail_sf
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR019749 Band_41_domain
IPR011259 ERM_C_dom
IPR035963 FERM_2
IPR041789 ERM_FERM_C
IPR018979 FERM_N
IPR019747 FERM_CS
IPR019748 FERM_central
IPR000798 Ez/rad/moesin-like
IPR011993 PH-like_dom_sf
IPR018980 FERM_PH-like_C
IPR000299 FERM_domain
IPR029071 Ubiquitin-like_domsf
IPR008954 Moesin_tail_sf
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR019749 Band_41_domain
IPR011259 ERM_C_dom
IPR035963 FERM_2
Gene 3D
ProteinModelPortal
H9JMY2
A0A2H1W6G9
A0A2W1BML9
A0A212F3B6
A0A194PXM7
A0A3S2NMJ8
+ More
D6WLL4 A0A1Y1NE13 N6UCS1 A0A0T6BFX0 E2BMZ0 A0A0L7RIW4 E0W3K1 A0A195CJU2 K7J0L8 A0A232ERE5 A0A1B6GIY0 A0A1B6DKY5 A0A1B6GCM0 A0A088AHV1 A0A0J7KWG0 A0A026WL04 F4WTL0 A0A3L8D6W2 A0A0A9XPR7 A0A158NGF8 A0A1B6MSV4 A0A0A9XLI6 A0A0K8SRW7 A0A195F1X6 A0A154PS47 E2A9J4 A0A151XBA2 A0A195E7A3 E9I939 A0A0C9QQB3 A0A0C9RC71 A0A0C9PWL1 A0A224XRV0 A0A151I145 A0A2I2Y252 Q8T898 H2XJZ5 A0A0N0BIU4 A0A3B4TER1 T1HAT2 A0A3B4YVG8 G3QMQ1 A0A267DEC2 A0A3B4ZH51 A0A1I8HW35 A0A2G8LDS9 A0A3Q1K2A6 A0A2I3NBN1 A0A2A3EA67 A0A2K5MR19 A0A2I3N1X3 A0A3Q3RR07 A0A2K6MRV6 A0A337SAG0 A0A0F8C8K1 I3JFD2 A0A2K6P9B0 A0A2K5ZYV5 A0A096N0X5 A0A2F0BJ09 A0A3Q1BH65 A0A091DRE1 B0CM43 A0A3P8S2Q7 M3ZBJ3 A0A091I0V5 A0A3B4H8N2 A0A3Q2VUW3 A0A3B4HB00 A0A3P9AUF2 B3RY22 A0A3P8R0P4 A0A369SLD1 A0A2K6D0C1 A0A2K5VVN2 F7GJB5 A0A2K6P9B7 A0A2I3M6Y6 G5BCM0 A0A3P8X401 A0A2Y9FBC9 A0A1U7SRI2 A0A3M6T605 Q4R807 A0A2R8ZKF1 A0A2I3T3D0 A0A0C4DG29 A0A2J8XAZ6 A0A2K5MR14 Q95KH8 A0A1D5Q281 A0A3Q4I1F9
D6WLL4 A0A1Y1NE13 N6UCS1 A0A0T6BFX0 E2BMZ0 A0A0L7RIW4 E0W3K1 A0A195CJU2 K7J0L8 A0A232ERE5 A0A1B6GIY0 A0A1B6DKY5 A0A1B6GCM0 A0A088AHV1 A0A0J7KWG0 A0A026WL04 F4WTL0 A0A3L8D6W2 A0A0A9XPR7 A0A158NGF8 A0A1B6MSV4 A0A0A9XLI6 A0A0K8SRW7 A0A195F1X6 A0A154PS47 E2A9J4 A0A151XBA2 A0A195E7A3 E9I939 A0A0C9QQB3 A0A0C9RC71 A0A0C9PWL1 A0A224XRV0 A0A151I145 A0A2I2Y252 Q8T898 H2XJZ5 A0A0N0BIU4 A0A3B4TER1 T1HAT2 A0A3B4YVG8 G3QMQ1 A0A267DEC2 A0A3B4ZH51 A0A1I8HW35 A0A2G8LDS9 A0A3Q1K2A6 A0A2I3NBN1 A0A2A3EA67 A0A2K5MR19 A0A2I3N1X3 A0A3Q3RR07 A0A2K6MRV6 A0A337SAG0 A0A0F8C8K1 I3JFD2 A0A2K6P9B0 A0A2K5ZYV5 A0A096N0X5 A0A2F0BJ09 A0A3Q1BH65 A0A091DRE1 B0CM43 A0A3P8S2Q7 M3ZBJ3 A0A091I0V5 A0A3B4H8N2 A0A3Q2VUW3 A0A3B4HB00 A0A3P9AUF2 B3RY22 A0A3P8R0P4 A0A369SLD1 A0A2K6D0C1 A0A2K5VVN2 F7GJB5 A0A2K6P9B7 A0A2I3M6Y6 G5BCM0 A0A3P8X401 A0A2Y9FBC9 A0A1U7SRI2 A0A3M6T605 Q4R807 A0A2R8ZKF1 A0A2I3T3D0 A0A0C4DG29 A0A2J8XAZ6 A0A2K5MR14 Q95KH8 A0A1D5Q281 A0A3Q4I1F9
Ontologies
PANTHER
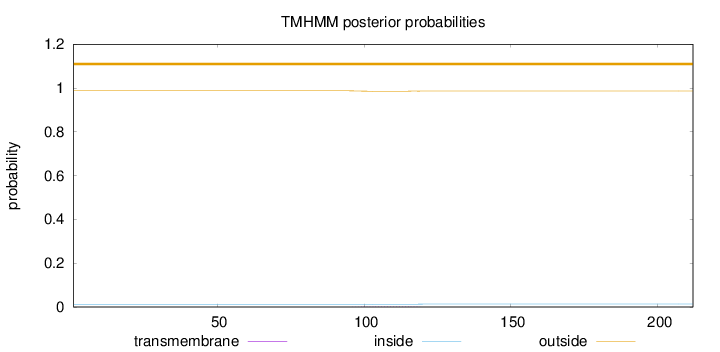
Topology
Length:
212
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0388
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01281
outside
1 - 212
Population Genetic Test Statistics
Pi
23.973262
Theta
25.264777
Tajima's D
-0.140902
CLR
4.209007
CSRT
0.342682865856707
Interpretation
Uncertain
