Pre Gene Modal
BGIBMGA010866
Annotation
PREDICTED:_uncharacterized_protein_HI_1014-like_[Amyelois_transitella]
Full name
D-erythronate dehydrogenase
Location in the cell
PlasmaMembrane Reliability : 1.278
Sequence
CDS
ATGAAAGTGGTCATAACAGGAGCTGCTGGCTTTTTGGGTTCGCGTTTAGCTGATGCTTTGCTAAAACCAAATTCCGAACTGTGTGTGACTGAATTGGTGTTAGTGGATACGATCTTACCACCCTCCCGAAAAGATCCACGAGTAATAACACTCGCCCTCGATATCACAACACCCGAGGCTGCGCGTAAACTCGTTGATACTGATACCGATGTACTGTTTCATTTGGCTGCAGTCGTAAGTGGACATGCTGAAGAAGACTTCGATCTTGGAATGAAAGTGAATTTCAACGCAACGAGATCATTGCTGGAGGCAGCGAGACATCGAGCTACGAAGCTTCGTTTTGTTTTTACCAGTACTGTTGGGGTGTTCGGAGGTGACCTGCCCCCTATTATAACTGATCAAACTGCCGCAACACCTCAGAACTCTTACGGCATTGCTAAGGCTATGAGCGAGCTTCTTATCAATGATTACTCTAGACGAGGTTTCGTTGATGGGAGAGTTGTTCGTCTTCCAACAGTTAGTATTCGCGCCGGAACTGCCAACAGAGCCGTGACATCTTTTGCAAGCGGAATTATCAGGGAGCCATTGAATGGTGAAGTAGCTATCTGCCCCGTTCCACCTGATCAAGAGTTATGGCTTTCAAGTCCTGGATCTGTTGTGTATAATTTAATACACGCTGCTACTTTACTTAGCTCCGAATTAGAGAAATGGAGAGTGATAAATCTACCTGGAATCAGTGTCACAGTTCGTAAAATGATTTCCTCGTTGTGTGAAGTTTCTGGAAGCAAGGTTGCTAAGCTGGTACGCTATGAACACGATGAGACCATAAGTGCAATGGTCGGCAGTTTCCCAAGCAGGTTCGACAACTCACGAGCATTGAGGCTTGGATTTCGAGTGGATTCTAATTTTACGGATGTGATACGGTCCTATATATTGGAAGATCTAAAACAGGTTAGCTTCGGTTTTTGA
Protein
MKVVITGAAGFLGSRLADALLKPNSELCVTELVLVDTILPPSRKDPRVITLALDITTPEAARKLVDTDTDVLFHLAAVVSGHAEEDFDLGMKVNFNATRSLLEAARHRATKLRFVFTSTVGVFGGDLPPIITDQTAATPQNSYGIAKAMSELLINDYSRRGFVDGRVVRLPTVSIRAGTANRAVTSFASGIIREPLNGEVAICPVPPDQELWLSSPGSVVYNLIHAATLLSSELEKWRVINLPGISVTVRKMISSLCEVSGSKVAKLVRYEHDETISAMVGSFPSRFDNSRALRLGFRVDSNFTDVIRSYILEDLKQVSFGF
Summary
Description
Catalyzes oxidation of D-erythronate to 2-oxo-tetronate. Can use either NAD(+) or NADP(+) as cosubstrate, with a preference for NAD(+).
Catalytic Activity
D-erythronate + NAD(+) = 2-dehydro-D-erythronate + H(+) + NADH
Biophysicochemical Properties
0.23 mM for NAD(+)
0.61 mM for NADP(+)
0.61 mM for NADP(+)
Similarity
Belongs to the NAD(P)-dependent epimerase/dehydratase family.
Keywords
Carbohydrate metabolism
Complete proteome
NAD
NADP
Oxidoreductase
Reference proteome
Feature
chain D-erythronate dehydrogenase
Uniprot
H9JMW3
A0A2A4K0H4
H9JL84
A0A2H1WVB3
A0A194REB2
A0A194PYH2
+ More
A0A212F134 A0A0L7L4Y4 C7BIG8 A0A2X5ELF4 D4DXV8 A0A0F7HA47 A0A3S5F1B9 A0A2V4FZ93 A0A2R3GWM3 A0A3F3KTC8 A0A329X2G6 A0A1C0U4Z5 A0A3S5DF99 A0A0U6JXX8 A0A126VBV8 L0MC25 D1P6T1 A0A329WP49 A0A329WIY6 A0A329WL77 A0A1G5RLF7 A0A379G080 A0A3G2FNV5 K8VZG1 A0A329XCE1 A0A1C0NQJ7 W3YBA5 A0A168I068 U2MBC8 A0A3R2DK46 Q7N442 A0A2S8QS65 A0A329VHS3 A0A1J0E352 K8VXV8 B6XDF6 A0A1C7WBC2 A0A291E7M3 X6QE30 A0A1S8CHY3 A0A081S1B7 A0A1Q4P2P9 A0A1B7JRJ0 T0P4M0 A0A292A9P0 A0A022PMD3 A0A1S1HR13 A0A370S2E9 A0A140NJZ1 A0A379HFI3 B2PVQ6 A0A335E9J2 A0A379GWD1 A0A1Q5VIB9 U7R030 D4BZD2 A0A3N1ZEI9 A0A1B8SX17 A0A254MIU1 A0A379FLP5 A0A3R8XBY2 A0A345LXR4 A0A2S8QSR2 A0A356D437 A0A0U4IAJ3 W1IV05 A0A2S8Q7N3 A0A068QYJ7 A0A2D0LCE0 A0A1B8YJN0 A0A2V1HC65 A0A2S8QGB1 A0A1I5CQ49 A0A0A0CVG1 A0A2G0Q4H6 A0A1Q5TVI1 W3V3V3 A0A1V3JTD1 B0UWX0 Q0I1A7 A0A1Y0IZ38 A0A1V4B0X9 A0A2X4XS93 A0A0Y0BN71 A0A0E1SQ33 A0A1V3KLI0 A0A2X4I057 A0A2S9RQZ9 A0A2S9RL90 P44094 A0A380TPX1 A4N1V8 A0A1J4QC82 A0A3E1QXW8
A0A212F134 A0A0L7L4Y4 C7BIG8 A0A2X5ELF4 D4DXV8 A0A0F7HA47 A0A3S5F1B9 A0A2V4FZ93 A0A2R3GWM3 A0A3F3KTC8 A0A329X2G6 A0A1C0U4Z5 A0A3S5DF99 A0A0U6JXX8 A0A126VBV8 L0MC25 D1P6T1 A0A329WP49 A0A329WIY6 A0A329WL77 A0A1G5RLF7 A0A379G080 A0A3G2FNV5 K8VZG1 A0A329XCE1 A0A1C0NQJ7 W3YBA5 A0A168I068 U2MBC8 A0A3R2DK46 Q7N442 A0A2S8QS65 A0A329VHS3 A0A1J0E352 K8VXV8 B6XDF6 A0A1C7WBC2 A0A291E7M3 X6QE30 A0A1S8CHY3 A0A081S1B7 A0A1Q4P2P9 A0A1B7JRJ0 T0P4M0 A0A292A9P0 A0A022PMD3 A0A1S1HR13 A0A370S2E9 A0A140NJZ1 A0A379HFI3 B2PVQ6 A0A335E9J2 A0A379GWD1 A0A1Q5VIB9 U7R030 D4BZD2 A0A3N1ZEI9 A0A1B8SX17 A0A254MIU1 A0A379FLP5 A0A3R8XBY2 A0A345LXR4 A0A2S8QSR2 A0A356D437 A0A0U4IAJ3 W1IV05 A0A2S8Q7N3 A0A068QYJ7 A0A2D0LCE0 A0A1B8YJN0 A0A2V1HC65 A0A2S8QGB1 A0A1I5CQ49 A0A0A0CVG1 A0A2G0Q4H6 A0A1Q5TVI1 W3V3V3 A0A1V3JTD1 B0UWX0 Q0I1A7 A0A1Y0IZ38 A0A1V4B0X9 A0A2X4XS93 A0A0Y0BN71 A0A0E1SQ33 A0A1V3KLI0 A0A2X4I057 A0A2S9RQZ9 A0A2S9RL90 P44094 A0A380TPX1 A4N1V8 A0A1J4QC82 A0A3E1QXW8
EC Number
1.1.1.410
Pubmed
EMBL
BABH01029054
NWSH01000283
PCG77775.1
BABH01011773
ODYU01011283
SOQ56922.1
+ More
KQ460323 KPJ15927.1 KQ459586 KPI98073.1 AGBW02010988 OWR47421.1 JTDY01002925 KOB70475.1 FM162591 CAQ84106.1 LS483492 SQJ30205.1 ADBY01000017 EFE97450.1 CP011254 AKG69622.1 LR134492 VEI63131.1 QJPU01000002 PYA07746.1 CP023268 AVJ16073.1 NSCN01000005 RAX11134.1 LOMY01000069 OCQ52997.1 LR134155 VEA71535.1 FCGE01000077 QJPV01000002 QMDQ01000001 CVA98068.1 PYA41669.1 RAU89417.1 CP003942 AGB80847.1 ABXV02000046 EFB70778.1 NSCL01000001 RAX06309.1 NSCJ01000001 RAX04691.1 NSCK01000001 RAX04360.1 FMWJ01000050 SCZ74221.1 UGUA01000002 SUC34419.1 CP033055 AYM93289.1 AKKL01000055 EKT52931.1 NSCM01000001 RAX14509.1 LJDX02000239 OCO18733.1 AZYZ01000045 ETS98960.1 FKZE01000007 SAQ07981.1 ASZB01000005 ERK14048.1 RCUM01001136 RCUN01000468 MNG78800.1 MNH81218.1 BX571867 CAE14886.1 PUWX01000015 PQQ38146.1 NSCI01000015 RAW90597.1 CP027418 AVL73910.1 AKKN01000014 EKT53073.1 ABXW01000019 EEB46654.1 LXKR01000022 OCJ38748.1 CP023536 ATG15145.1 JACS01000029 ETT06604.1 MOXD01000008 OMQ21308.1 JGVH01000004 KER04720.1 MJAO01000006 OKB67394.1 LXEW01000037 OAT50516.1 AUXQ01000051 EQB99291.1 CP023956 ATM75617.1 JFGV01000014 EYU16133.1 LVIE01000163 OHT24272.1 QRAU01000031 RDL13920.1 CP003488 AFH93425.1 UGUC01000003 SUC77871.1 ABJD02000048 EDU61443.1 UFMN01000011 SST04674.1 UGUB01000002 SUC45221.1 MQRH01000003 OKP30420.1 AXDT01000145 ERT12156.1 ACCI02000050 EFE53506.1 RJVN01000019 ROR48497.1 LYBX01000002 NOWC01000008 NXKD01000030 QUAF01000008 UAUG01000013 OBY37303.1 OZS74922.1 PCQ36096.1 RFT08249.1 SPZ21381.1 MYFJ01000048 OWO80501.1 UGTZ01000001 SUC29724.1 RHRR01000009 RRW33366.1 CP031123 AXH62904.1 PUWW01000144 PQQ38239.1 DOEH01000012 HBO22375.1 CP013913 ALX93215.1 CBXF010000057 CDL81673.1 PUWT01000007 PQQ28851.1 FO704550 CDG18900.1 NJCX01000012 PHM73358.1 LOIC01000042 OCA55298.1 PYUJ01000048 PVZ82863.1 PUWU01000009 PQQ34307.1 FOVO01000029 SFN89135.1 JQOC01000001 KGM29618.1 NJAI01000005 PHM54120.1 MKGR01000022 OKP04237.1 AYSJ01000013 ETS30596.1 MLHQ01000006 OOF59978.1 CP000947 ACA31698.1 CP000436 ABI24297.1 CP018803 ARU65821.1 UGHF01000001 STO59451.1 LS483480 SQI78949.1 FCYU01000003 CVP60354.1 AAZI01000001 EDK12402.1 MLAE01000023 OOF78521.1 LS483411 SQG88488.1 NEBY01000142 LS483392 LR134490 PRJ63462.1 SQG36978.1 VEI56924.1 NEBH01000014 PRJ32470.1 L42023 UFRQ01000003 SUT88921.1 AAZE01000001 EDJ91816.1 MDKE01000055 OIN05547.1 QVIZ01000004 RFN95431.1
KQ460323 KPJ15927.1 KQ459586 KPI98073.1 AGBW02010988 OWR47421.1 JTDY01002925 KOB70475.1 FM162591 CAQ84106.1 LS483492 SQJ30205.1 ADBY01000017 EFE97450.1 CP011254 AKG69622.1 LR134492 VEI63131.1 QJPU01000002 PYA07746.1 CP023268 AVJ16073.1 NSCN01000005 RAX11134.1 LOMY01000069 OCQ52997.1 LR134155 VEA71535.1 FCGE01000077 QJPV01000002 QMDQ01000001 CVA98068.1 PYA41669.1 RAU89417.1 CP003942 AGB80847.1 ABXV02000046 EFB70778.1 NSCL01000001 RAX06309.1 NSCJ01000001 RAX04691.1 NSCK01000001 RAX04360.1 FMWJ01000050 SCZ74221.1 UGUA01000002 SUC34419.1 CP033055 AYM93289.1 AKKL01000055 EKT52931.1 NSCM01000001 RAX14509.1 LJDX02000239 OCO18733.1 AZYZ01000045 ETS98960.1 FKZE01000007 SAQ07981.1 ASZB01000005 ERK14048.1 RCUM01001136 RCUN01000468 MNG78800.1 MNH81218.1 BX571867 CAE14886.1 PUWX01000015 PQQ38146.1 NSCI01000015 RAW90597.1 CP027418 AVL73910.1 AKKN01000014 EKT53073.1 ABXW01000019 EEB46654.1 LXKR01000022 OCJ38748.1 CP023536 ATG15145.1 JACS01000029 ETT06604.1 MOXD01000008 OMQ21308.1 JGVH01000004 KER04720.1 MJAO01000006 OKB67394.1 LXEW01000037 OAT50516.1 AUXQ01000051 EQB99291.1 CP023956 ATM75617.1 JFGV01000014 EYU16133.1 LVIE01000163 OHT24272.1 QRAU01000031 RDL13920.1 CP003488 AFH93425.1 UGUC01000003 SUC77871.1 ABJD02000048 EDU61443.1 UFMN01000011 SST04674.1 UGUB01000002 SUC45221.1 MQRH01000003 OKP30420.1 AXDT01000145 ERT12156.1 ACCI02000050 EFE53506.1 RJVN01000019 ROR48497.1 LYBX01000002 NOWC01000008 NXKD01000030 QUAF01000008 UAUG01000013 OBY37303.1 OZS74922.1 PCQ36096.1 RFT08249.1 SPZ21381.1 MYFJ01000048 OWO80501.1 UGTZ01000001 SUC29724.1 RHRR01000009 RRW33366.1 CP031123 AXH62904.1 PUWW01000144 PQQ38239.1 DOEH01000012 HBO22375.1 CP013913 ALX93215.1 CBXF010000057 CDL81673.1 PUWT01000007 PQQ28851.1 FO704550 CDG18900.1 NJCX01000012 PHM73358.1 LOIC01000042 OCA55298.1 PYUJ01000048 PVZ82863.1 PUWU01000009 PQQ34307.1 FOVO01000029 SFN89135.1 JQOC01000001 KGM29618.1 NJAI01000005 PHM54120.1 MKGR01000022 OKP04237.1 AYSJ01000013 ETS30596.1 MLHQ01000006 OOF59978.1 CP000947 ACA31698.1 CP000436 ABI24297.1 CP018803 ARU65821.1 UGHF01000001 STO59451.1 LS483480 SQI78949.1 FCYU01000003 CVP60354.1 AAZI01000001 EDK12402.1 MLAE01000023 OOF78521.1 LS483411 SQG88488.1 NEBY01000142 LS483392 LR134490 PRJ63462.1 SQG36978.1 VEI56924.1 NEBH01000014 PRJ32470.1 L42023 UFRQ01000003 SUT88921.1 AAZE01000001 EDJ91816.1 MDKE01000055 OIN05547.1 QVIZ01000004 RFN95431.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000002747 UP000249094 UP000005723 UP000034699 UP000270487 UP000248086 UP000238421 UP000251048 UP000093476 UP000271603 UP000054899 UP000247670 UP000251655 UP000011001 UP000005512 UP000251134 UP000251359 UP000251400 UP000183223 UP000255129 UP000281837 UP000009336 UP000250919 UP000050507 UP000019057 UP000078180 UP000016623 UP000002514 UP000250870 UP000239526 UP000010290 UP000003729 UP000093001 UP000217606 UP000022577 UP000216021 UP000028002 UP000185770 UP000078224 UP000015813 UP000221800 UP000023464 UP000179588 UP000255042 UP000005012 UP000254690 UP000004506 UP000262355 UP000254419 UP000186436 UP000017133 UP000003321 UP000273646 UP000216001 UP000218329 UP000250984 UP000260960 UP000197563 UP000254208 UP000275009 UP000256296 UP000239084 UP000263856 UP000061771 UP000019202 UP000239550 UP000032721 UP000221101 UP000092665 UP000244888 UP000239784 UP000199011 UP000029997 UP000225433 UP000186277 UP000018957 UP000188602 UP000008543 UP000001970 UP000195281 UP000254329 UP000248560 UP000248527 UP000005706 UP000189114 UP000249265 UP000238532 UP000249751 UP000278034 UP000237959 UP000000579 UP000254649 UP000003798 UP000243073 UP000261070
UP000002747 UP000249094 UP000005723 UP000034699 UP000270487 UP000248086 UP000238421 UP000251048 UP000093476 UP000271603 UP000054899 UP000247670 UP000251655 UP000011001 UP000005512 UP000251134 UP000251359 UP000251400 UP000183223 UP000255129 UP000281837 UP000009336 UP000250919 UP000050507 UP000019057 UP000078180 UP000016623 UP000002514 UP000250870 UP000239526 UP000010290 UP000003729 UP000093001 UP000217606 UP000022577 UP000216021 UP000028002 UP000185770 UP000078224 UP000015813 UP000221800 UP000023464 UP000179588 UP000255042 UP000005012 UP000254690 UP000004506 UP000262355 UP000254419 UP000186436 UP000017133 UP000003321 UP000273646 UP000216001 UP000218329 UP000250984 UP000260960 UP000197563 UP000254208 UP000275009 UP000256296 UP000239084 UP000263856 UP000061771 UP000019202 UP000239550 UP000032721 UP000221101 UP000092665 UP000244888 UP000239784 UP000199011 UP000029997 UP000225433 UP000186277 UP000018957 UP000188602 UP000008543 UP000001970 UP000195281 UP000254329 UP000248560 UP000248527 UP000005706 UP000189114 UP000249265 UP000238532 UP000249751 UP000278034 UP000237959 UP000000579 UP000254649 UP000003798 UP000243073 UP000261070
Pfam
PF01370 Epimerase
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JMW3
A0A2A4K0H4
H9JL84
A0A2H1WVB3
A0A194REB2
A0A194PYH2
+ More
A0A212F134 A0A0L7L4Y4 C7BIG8 A0A2X5ELF4 D4DXV8 A0A0F7HA47 A0A3S5F1B9 A0A2V4FZ93 A0A2R3GWM3 A0A3F3KTC8 A0A329X2G6 A0A1C0U4Z5 A0A3S5DF99 A0A0U6JXX8 A0A126VBV8 L0MC25 D1P6T1 A0A329WP49 A0A329WIY6 A0A329WL77 A0A1G5RLF7 A0A379G080 A0A3G2FNV5 K8VZG1 A0A329XCE1 A0A1C0NQJ7 W3YBA5 A0A168I068 U2MBC8 A0A3R2DK46 Q7N442 A0A2S8QS65 A0A329VHS3 A0A1J0E352 K8VXV8 B6XDF6 A0A1C7WBC2 A0A291E7M3 X6QE30 A0A1S8CHY3 A0A081S1B7 A0A1Q4P2P9 A0A1B7JRJ0 T0P4M0 A0A292A9P0 A0A022PMD3 A0A1S1HR13 A0A370S2E9 A0A140NJZ1 A0A379HFI3 B2PVQ6 A0A335E9J2 A0A379GWD1 A0A1Q5VIB9 U7R030 D4BZD2 A0A3N1ZEI9 A0A1B8SX17 A0A254MIU1 A0A379FLP5 A0A3R8XBY2 A0A345LXR4 A0A2S8QSR2 A0A356D437 A0A0U4IAJ3 W1IV05 A0A2S8Q7N3 A0A068QYJ7 A0A2D0LCE0 A0A1B8YJN0 A0A2V1HC65 A0A2S8QGB1 A0A1I5CQ49 A0A0A0CVG1 A0A2G0Q4H6 A0A1Q5TVI1 W3V3V3 A0A1V3JTD1 B0UWX0 Q0I1A7 A0A1Y0IZ38 A0A1V4B0X9 A0A2X4XS93 A0A0Y0BN71 A0A0E1SQ33 A0A1V3KLI0 A0A2X4I057 A0A2S9RQZ9 A0A2S9RL90 P44094 A0A380TPX1 A4N1V8 A0A1J4QC82 A0A3E1QXW8
A0A212F134 A0A0L7L4Y4 C7BIG8 A0A2X5ELF4 D4DXV8 A0A0F7HA47 A0A3S5F1B9 A0A2V4FZ93 A0A2R3GWM3 A0A3F3KTC8 A0A329X2G6 A0A1C0U4Z5 A0A3S5DF99 A0A0U6JXX8 A0A126VBV8 L0MC25 D1P6T1 A0A329WP49 A0A329WIY6 A0A329WL77 A0A1G5RLF7 A0A379G080 A0A3G2FNV5 K8VZG1 A0A329XCE1 A0A1C0NQJ7 W3YBA5 A0A168I068 U2MBC8 A0A3R2DK46 Q7N442 A0A2S8QS65 A0A329VHS3 A0A1J0E352 K8VXV8 B6XDF6 A0A1C7WBC2 A0A291E7M3 X6QE30 A0A1S8CHY3 A0A081S1B7 A0A1Q4P2P9 A0A1B7JRJ0 T0P4M0 A0A292A9P0 A0A022PMD3 A0A1S1HR13 A0A370S2E9 A0A140NJZ1 A0A379HFI3 B2PVQ6 A0A335E9J2 A0A379GWD1 A0A1Q5VIB9 U7R030 D4BZD2 A0A3N1ZEI9 A0A1B8SX17 A0A254MIU1 A0A379FLP5 A0A3R8XBY2 A0A345LXR4 A0A2S8QSR2 A0A356D437 A0A0U4IAJ3 W1IV05 A0A2S8Q7N3 A0A068QYJ7 A0A2D0LCE0 A0A1B8YJN0 A0A2V1HC65 A0A2S8QGB1 A0A1I5CQ49 A0A0A0CVG1 A0A2G0Q4H6 A0A1Q5TVI1 W3V3V3 A0A1V3JTD1 B0UWX0 Q0I1A7 A0A1Y0IZ38 A0A1V4B0X9 A0A2X4XS93 A0A0Y0BN71 A0A0E1SQ33 A0A1V3KLI0 A0A2X4I057 A0A2S9RQZ9 A0A2S9RL90 P44094 A0A380TPX1 A4N1V8 A0A1J4QC82 A0A3E1QXW8
PDB
2HRZ
E-value=1.72637e-61,
Score=597
Ontologies
GO
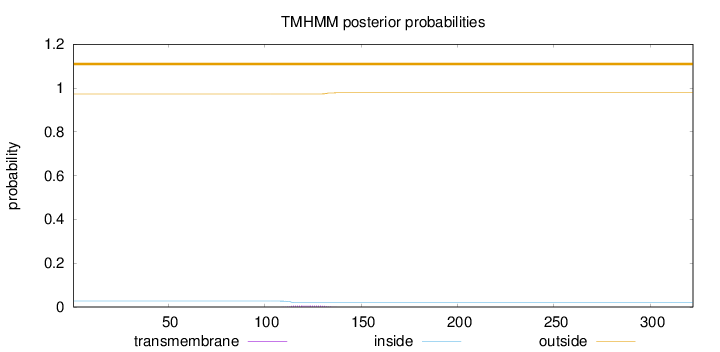
Topology
Length:
322
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12484
Exp number, first 60 AAs:
0.00227
Total prob of N-in:
0.02720
outside
1 - 322
Population Genetic Test Statistics
Pi
18.2039
Theta
18.915094
Tajima's D
-0.240706
CLR
2.111558
CSRT
0.312884355782211
Interpretation
Uncertain
