Pre Gene Modal
BGIBMGA010888
Annotation
PREDICTED:_glycogen_debranching_enzyme_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.163 Nuclear Reliability : 1.019
Sequence
CDS
ATGGCGATCGAGCGCAGCGCTGCCGCGGAGGCCGCGCGTGCCCCCGCCACGCCCACCGCAGCCCCCCTGCCCGCCCCCACCACCGTCATCACCCTCGAACACGGGGAGCATTTTGACTCTACGTTATATAGATTTGAAAAAGGCTGCTATTTACAATTTTGTCCCGGACCAAGTTTATTAGGCCGGAAGGTGTTCCTTTACACTAATTATGTGCTCACTGATGATGAACAAGACGATCAATCCGAGTTCATCCGCAATCAGTACTACGGGCTGGAGTGGCGCGCCGAGGGGGAGGAGGGAGAGAAGGGCGAGTCGTTGGGAGCCGGGCCCCTCGTCACCGACACCGACCTCTACTGTAAGATCAAGCTGGCCAGGGCTGGCTCCTTCCATTTCTACTTCGTCTATGATAATGCAGAATCTAAGATAGGTCCCCAAGGCTCGGGCTGGTTCCACGTGGCGCCGACGCTGCGCGCGGGCCCGGGCGGCGCGGCGCGCATCCCGCTGGACGGCGTCGCCATGCAGACCGTGCTGGCCAAGAGCCTGGGCCCCTTGCCGCGCTGGGAGAACACGCTCAGGGTCGCCAAAGAGACGGGCTATAATATGATACATTTTACTCCTATACAGGAGCTGGGCGGCTCCAACTCGAGCTACAGCATCGCCAACCAGCTGAAGCTGAACCCTCGCTTCGGGCAGGACACGGGGCGGGAGGTCACCTTCGCTGACGTCGAGAACATTGTCGCCAAGATCAAAAACGACTGGAACATGCTGTCGATATGCGACGTGGTGCTGAACCACACGGCCAACGAGACGAGCTGGGTGGCGGAGCACCCCGAGGCCACGTACAACTGCGGCTCGTGCGCCTACCTGCGCCCCGCCGCGCTGCTGGACGCCGCGCTCGCCGCCCTCACCGCGCGCGTCGCCCGCGCCCTGCACGCGCCGCCCGTGCCGCCGCGCGTCACGCAACACGCGCACGTCGAGGCCATCAAACACATATTGGAGACGCAGGTAGTCCCAGAGCTGCGCCTGCACGAAATGTACATGTGCAATGTGTCGGAGCTAGTCCAGGAGTTCTACTTTTTGGCGAGAAACAAGGTGCCGGCGGTGAAGGCCCCGGCGGAGGCGGAGCGGCCGGTGCTGAAGCTGGTGGAGGACCCGCAGTACCGCCGCCTCCGCGCCACCGTCGACCTGGACGCCGCTCTGCAGCTCTACAACGTGTACAGGTCGGAGTGTTACGACGAGGAGTCCCGCCTCAAGAAATGCTGTGAGGAGTTGAAGCTGAACCTGGAGCAGCTGAACGAGGCGGCCGCGGAGCAGGTGCGGGGACACCTCCGCGCCGCCGTCGACAACGTGGTCGCCGGCATGAGGTACTACCGCCTGCAATCGGACGGACCCAAGATCGAGGAAGTCAGCGCTAAGAATCCGCTGGTGCCGAGGTACTTCACGTGGGCGTGCGACGTGGAGTCGTGCAGCAGCGCGGAGGTGGAGGCGGCGCTGTACGGGGGGGAGGGGGGCGCGGTGATGGCGCACAACGGCTGGGTCATGGGGGCCGACCCCCTCACGGACTTCGCCGCGCGCCCCGCCGCCGCCGTCTACCTGCGCCGTGACCTGCTGGCCTGGGGGGACTCCATCAAGCTCAGGTACGGCGACAAGCCGGAGGACAGCCCGTTCCTGTGGGCGCACATGCGGGAGTACGTGGAGCTCACGGCCGAAGTGTTCGACGGACTGCGCCTCGACAACTGCCACTCCACGCCGCTGCACGTGGCGGAGTACATGCTGGACTGCGCGCGCAACGTCAAGCCGGACCTGTACGTGGTGGCGGAACTCTTCACCAACTCCGACCACGTTGACAACATCTTCGTCAACCGACTGGGCATCACTTCTCTCATCAGAGAGGCTCAGAGCGCGTGGGACTCGCACGAGCAGGGCCGGCTCCTGCACCGGTTCGGGGGGCGCGGCGCGGGGGCCTTCATGTGGGGGGGGCCGCGCCCCGCCGTGCCGCACGTGGCGCGCGCCGCGCTCTACGACCTCACGCACGACAACCCCTCGCCCATCGACAAGAGATCCGTCTGGGATTTGTTGCCGTCGTCGGCCTTGGTGTCCATGGCCTGCTGCGCCACGGGGAGCACGCGCGGCTACGACGAGCTGGTCCCCCACCACGTCCACGTGGTCGACGAGTCGCGGCTGTACTGCGAGTGGAGCGAGGGCGCGGAGCCGGCCGAGGGCGGCGTGGGCCCCGCCTGCGGGATCCTGGCCGCCAAGCGAGCCCTCAACGAGCTGCACCTACAGCTGGCCCAGGGCGGCTACGACGAGGTGTACGTGGACCAGATGGACGCTGACGTGGTGGCCGTGACGCGCCACGAGCCGCGCTCCCGCCGCTCCGTCATCGCCGTCGCCTTCACCGCCTTCCGCCCCCCCGCCGCCGCCGCCCGCCGCGCGCCCCCACCCCTCCGCTTTGAGGGACAGCTCGAGCAGATCCTCATCGAAACTGGATTGAGAATCAAGGACCACACGGCCGGCCCGTTCTCGAAGCACCCCCGCTACATCACCGGGCTCGCGCAGTACGACGCGCCCGTGCGCCGCGACGTGCCGCTCGCGCAGTCCGACGTGTTCGCCGCGGAGAGGACCGAAGGACAAACCACGATCTTGGAATTCAAGCCCTTATCCCCGGGCACTGTGGTCGTCATTCGCGTGGCGCCGCGCCCGGCCGCCGCCAGCGCCGTGGCCGCGCTGCACAGGCACATGGAGCTGGCGGGCGGCGGCGACCCGTACGGCCTGCAGCCGGCCCTGGCCGAGCTCGACCTGGTGGACTTCAACGTGCTGCTGTACAGATGCGACGCGGAGGAGAGGGACGCGGGGGGCGGCGGAGTCTACGACGTGCCGGGCCACGGGCCCCTCGTGTACGCCGGCCTGCAGGGCGTGATCTCGTTACTCTCGGAGATAACTCCCAACGACGACCTGGGTCACCCGCTGTGCGACAACCTGCGCGCCGGGGACTGGCTCATGGACTACACGTGCGCGCGCCTGGAGAGGGACCCCAGACTGGCTGCCGTGGCCGCCAAGTACAGGGAGGCGTTCCGGCCCATCTCTGCGCTGCCCCGGTTCCTGGTGCCGGCGTACTTCGCGGCGGCGGCGGGTGCGCTGCACGGCGCGCTGGTGGCGGCGGCGCTGCGGCGGCTGCGCGCGCCCGCGGGGACCCTGCGCGCCGCGCTCGCGCTCACCAGCGTGCAGCTCACAGGGCACGTGCCGAGCGCGACCCTGCCCGCGCCGCTCCCCGCGCCCCCCACCGCGCCGGCCCCCCGCGGCCCCCGCCCCGGCTCGCTGTCCGCCGGCCTGCCGCACTTCGTGGTGGGCTACACGCGCTGCTGGGGGCGCGACACCTTCATAGCGCTCCGCGGCATGTTCCTGCTCACCGGGAGATACGAGGACGCTCGCCTCCACATACTGTCGTACGGGGCGTGTCTCCGGCACGGCCTGATCCCCAACCTGCTGGACTGCGGGAAGAACGCGCGCTACAACTGCCGCGACGCCGTGTGGTGGTGGCTCTACTGCGTGCAGCAGTATTGTAAGGAGGCCCCGGACGGGCTGTCCCTCCTGTCGGAGCCGGTGTGTCGGCTGCAGGCGCGCGAGGAGGGCGCCCCCGCCGCCGCGCCGCTGCACGAGCTGCTGCAGGACGCGCTCGACACGCACTTCCAGGGCCTCGTGTTCCGCGAGAGGAACGCCGGCAGGACCATCGACGCGCACATGACCGACAAGGGTTTTAATGTGCAAATCGGCATCGACCCAGAAACCGGGTTCCCGTTCGGTGGCAACGACTCGAATTGCGGCACGTGGATGGACAAGATGGGGTCGTCCGACAAGGCGGGCAACCGCGGACAGCCCGCCACCCCCCGGGACGGCAGCGCCATCGAGCTGGTCGCGCTGGCCTACAGCACCGCGCGCTGGCTGGCCGAGCTGCACCGCGCCGGGCGCTACCCGACCCCGGGGCTGGTGCGCCGCCACCGGGACGGCACGCTCACCTCCTGGACCTTCGCGCAGTGGGCCGACCGCATCCGGGCCTCCTTCGAGCGCCACTTCTGGGTCCCGCCGACCCCCGCCGCGCCCGACGCGCGCCCCGACCTGGTGCACCGCCGCGCCATCTACAAGGACACGCACGGCGCCTCGCGGCCCTGGGCCGACTACCAGCTGCGTTGCAACTTCCCCGTGGCCATGGCGCTCGCCCCCGACCTGTTCGACCCGCGCCACGCCTGGCAGGCCCTGGATCAGGTCGAGAAGCTGCTGCTGGGGCCGCTCGGCGTGAAGACGCTGGACCCGGCCGACTGGGCCTACCGACCGGACTACGACAACTCCAACGACTCGGACGACCCGAGCGTCGCGCACGGCTTCAACTACCACCAGGGCCCCGAGTGGGGCTGGCCGCTCGGCTTCTACGTGCGCGCGCGCCTGGCCTTCGCGCACCCCTGCGGCAAGCACGCCCGCGCCGTCGCCGCCGGGGCCCGGCCGGCGGCGCGCACCATCTACAACCTGCTGCAGGGCGGCCGCGACAGGCCGCTCTCTACCGTCGTGTACAGCGACCGGCTGCTGCGCTACGGGCTGCCGCTGCCGGTGCGCGTGGGGCGGCGCCGCCTGTGGGACGAGCTCGGCCGCGGCGCCGTCACGGAGCGTCGCGCCGCCGGGCCCCGCGCCGCGCACGTCCTGTCCCTCCCGCCGGTGCCGCGACTCGCCCCCCGCGCCGGGCCCCTGCTGGGTATCCCTTCTCCCAACGATTGTACAAAGTATTATCCGCGACGACAATTTCGATTTAGATCTTCTCGAAGACGAATCTTCATCCTGCCGGCCGATTGTCATTTCTTGGAATCGATCCTGTGGGAGGGCGAATCGGTTCACCGCTCTCTTGACTACCGGCAGCCCGCGGGTGGCGTCGAGCCCTGCCGCCCGCGCCCGGTCGCTCTGAAGCGGCTCTCTCGGATGTCTTCGGCCTGCGGCTCCGCGCTGGAGGAGCCGGACGAAGGGGGGATCCGGCCCCAGTTCGCGATGCCGGACGACGAGAGGGCGGCGGGCGGGGCGGGCGGGGAGGAGGAGGGGAGCGAGGGGGAGGCGGCGGAGCTGACGTACGAGGCGCTAGTGAAGCCCTCCCCGCGGGTCAGCGTCATCAATTTGTCGGAGCGGCAGAACAACAAGGCGGCGTGGCGGCCGCTGGAGCGCAAGGTCCTGAACACGGCGCGCGAGGGGGCGGGGGCGGGGGGGCGCGGGGCGCGGCTGCAGCGGGCCACGCTGCGCGCGCTGCTCTCCTTCTCGGGGCTCTTCTACACGGTCGCCTTCCTCACCTTCTACTTCCTGAGCTTGCCGTGA
Protein
MAIERSAAAEAARAPATPTAAPLPAPTTVITLEHGEHFDSTLYRFEKGCYLQFCPGPSLLGRKVFLYTNYVLTDDEQDDQSEFIRNQYYGLEWRAEGEEGEKGESLGAGPLVTDTDLYCKIKLARAGSFHFYFVYDNAESKIGPQGSGWFHVAPTLRAGPGGAARIPLDGVAMQTVLAKSLGPLPRWENTLRVAKETGYNMIHFTPIQELGGSNSSYSIANQLKLNPRFGQDTGREVTFADVENIVAKIKNDWNMLSICDVVLNHTANETSWVAEHPEATYNCGSCAYLRPAALLDAALAALTARVARALHAPPVPPRVTQHAHVEAIKHILETQVVPELRLHEMYMCNVSELVQEFYFLARNKVPAVKAPAEAERPVLKLVEDPQYRRLRATVDLDAALQLYNVYRSECYDEESRLKKCCEELKLNLEQLNEAAAEQVRGHLRAAVDNVVAGMRYYRLQSDGPKIEEVSAKNPLVPRYFTWACDVESCSSAEVEAALYGGEGGAVMAHNGWVMGADPLTDFAARPAAAVYLRRDLLAWGDSIKLRYGDKPEDSPFLWAHMREYVELTAEVFDGLRLDNCHSTPLHVAEYMLDCARNVKPDLYVVAELFTNSDHVDNIFVNRLGITSLIREAQSAWDSHEQGRLLHRFGGRGAGAFMWGGPRPAVPHVARAALYDLTHDNPSPIDKRSVWDLLPSSALVSMACCATGSTRGYDELVPHHVHVVDESRLYCEWSEGAEPAEGGVGPACGILAAKRALNELHLQLAQGGYDEVYVDQMDADVVAVTRHEPRSRRSVIAVAFTAFRPPAAAARRAPPPLRFEGQLEQILIETGLRIKDHTAGPFSKHPRYITGLAQYDAPVRRDVPLAQSDVFAAERTEGQTTILEFKPLSPGTVVVIRVAPRPAAASAVAALHRHMELAGGGDPYGLQPALAELDLVDFNVLLYRCDAEERDAGGGGVYDVPGHGPLVYAGLQGVISLLSEITPNDDLGHPLCDNLRAGDWLMDYTCARLERDPRLAAVAAKYREAFRPISALPRFLVPAYFAAAAGALHGALVAAALRRLRAPAGTLRAALALTSVQLTGHVPSATLPAPLPAPPTAPAPRGPRPGSLSAGLPHFVVGYTRCWGRDTFIALRGMFLLTGRYEDARLHILSYGACLRHGLIPNLLDCGKNARYNCRDAVWWWLYCVQQYCKEAPDGLSLLSEPVCRLQAREEGAPAAAPLHELLQDALDTHFQGLVFRERNAGRTIDAHMTDKGFNVQIGIDPETGFPFGGNDSNCGTWMDKMGSSDKAGNRGQPATPRDGSAIELVALAYSTARWLAELHRAGRYPTPGLVRRHRDGTLTSWTFAQWADRIRASFERHFWVPPTPAAPDARPDLVHRRAIYKDTHGASRPWADYQLRCNFPVAMALAPDLFDPRHAWQALDQVEKLLLGPLGVKTLDPADWAYRPDYDNSNDSDDPSVAHGFNYHQGPEWGWPLGFYVRARLAFAHPCGKHARAVAAGARPAARTIYNLLQGGRDRPLSTVVYSDRLLRYGLPLPVRVGRRRLWDELGRGAVTERRAAGPRAAHVLSLPPVPRLAPRAGPLLGIPSPNDCTKYYPRRQFRFRSSRRRIFILPADCHFLESILWEGESVHRSLDYRQPAGGVEPCRPRPVALKRLSRMSSACGSALEEPDEGGIRPQFAMPDDERAAGGAGGEEEGSEGEAAELTYEALVKPSPRVSVINLSERQNNKAAWRPLERKVLNTAREGAGAGGRGARLQRATLRALLSFSGLFYTVAFLTFYFLSLP
Summary
Uniprot
A0A2H1WHD1
A0A194PXK2
A0A1W5YN02
A0A0L7QQA0
A0A088ALS8
A0A154PFZ0
+ More
A0A2A3E3Q0 A0A158NS37 A0A195FW52 U4UE49 N6U2Q0 E2BPT9 A0A3L8DBU5 A0A026W7M0 A0A336MSF1 A0A336MMQ1 A0A336MYR3 A0A0M9A074 A0A0P5BVX3 A0A0P5BV94 A0A0P5ULF6 A0A0N7ZYU7 A0A0P5XGQ3 A0A0P5TY23 A0A0P5WNZ0 A0A0P6JIF6 A0A0P5A0R2 A0A0P5AGJ5 A0A0N7ZXC2 A0A0P4XYG0 A0A0P5ZAJ1 A0A0N8E1W1 A0A0P5XH45 A0A0P6C7D8 A0A0P5WLK6 A0A164NPU1 A0A0P5TL70 A0A0P5Z581 R7UWR3 A0A0P4X136 A0A0P5HQU5 V4ALP1 A0A0P5WIG7 A0A0P5WLX8 A0A3Q3XM66 A0A0P5TKV9 A0A0P5JL96 A0A0P5XPT4 A0A0P5NL68 A0A0N8EAH7 A0A0P5NYQ2 A0A0P5UWS3 A0A0P5SET8 A0A0P5QE56 A0A0P5UV70 A0A0P5Z6R4 A0A0P5DYV0 A0A0P5INE5 A0A0P5NN30 A0A0P5TT68 A0A0N8C7A5 A0A0R3PR76 A0A0P5B2G2 A0A0P5R4J9 A0A0N5AQW6 A0A0P5J4V2 F1KRD3 A0A0P6CYY7 E3MHZ4 A0A0N8CC17 A0A261BHA1 A0A0P5EPF5
A0A2A3E3Q0 A0A158NS37 A0A195FW52 U4UE49 N6U2Q0 E2BPT9 A0A3L8DBU5 A0A026W7M0 A0A336MSF1 A0A336MMQ1 A0A336MYR3 A0A0M9A074 A0A0P5BVX3 A0A0P5BV94 A0A0P5ULF6 A0A0N7ZYU7 A0A0P5XGQ3 A0A0P5TY23 A0A0P5WNZ0 A0A0P6JIF6 A0A0P5A0R2 A0A0P5AGJ5 A0A0N7ZXC2 A0A0P4XYG0 A0A0P5ZAJ1 A0A0N8E1W1 A0A0P5XH45 A0A0P6C7D8 A0A0P5WLK6 A0A164NPU1 A0A0P5TL70 A0A0P5Z581 R7UWR3 A0A0P4X136 A0A0P5HQU5 V4ALP1 A0A0P5WIG7 A0A0P5WLX8 A0A3Q3XM66 A0A0P5TKV9 A0A0P5JL96 A0A0P5XPT4 A0A0P5NL68 A0A0N8EAH7 A0A0P5NYQ2 A0A0P5UWS3 A0A0P5SET8 A0A0P5QE56 A0A0P5UV70 A0A0P5Z6R4 A0A0P5DYV0 A0A0P5INE5 A0A0P5NN30 A0A0P5TT68 A0A0N8C7A5 A0A0R3PR76 A0A0P5B2G2 A0A0P5R4J9 A0A0N5AQW6 A0A0P5J4V2 F1KRD3 A0A0P6CYY7 E3MHZ4 A0A0N8CC17 A0A261BHA1 A0A0P5EPF5
EMBL
ODYU01008617
SOQ52356.1
KQ459586
KPI98071.1
KX147680
ARI45581.1
+ More
KQ414786 KOC60813.1 KQ434896 KZC10742.1 KZ288391 PBC26357.1 ADTU01024457 ADTU01024458 KQ981208 KYN44656.1 KB632100 ERL88846.1 APGK01041757 KB740997 ENN75845.1 GL449658 EFN82290.1 QOIP01000010 RLU17593.1 KK107374 EZA51596.1 UFQT01001761 SSX31693.1 SSX31692.1 UFQT01002921 SSX34289.1 KQ435789 KOX74487.1 GDIP01179215 JAJ44187.1 GDIP01194666 JAJ28736.1 GDIP01201817 GDIP01111418 GDIP01044134 JAL92296.1 GDIP01199161 JAJ24241.1 GDIP01072629 JAM31086.1 GDIP01122822 JAL80892.1 GDIP01084523 JAM19192.1 GDIQ01007066 JAN87671.1 GDIP01205511 JAJ17891.1 GDIP01199162 JAJ24240.1 GDIP01203361 JAJ20041.1 GDIP01235107 JAI88294.1 GDIP01046881 JAM56834.1 GDIQ01069980 JAN24757.1 GDIP01084525 JAM19190.1 GDIP01012168 JAM91547.1 GDIP01084524 JAM19191.1 LRGB01002849 KZS06140.1 GDIP01124820 JAL78894.1 GDIP01048578 JAM55137.1 AMQN01005970 KB297235 ELU10759.1 GDIP01251954 JAI71447.1 GDIQ01224532 JAK27193.1 KB201802 ESO94506.1 GDIP01086026 JAM17689.1 GDIP01084526 JAM19189.1 GDIP01130323 JAL73391.1 GDIQ01203334 JAK48391.1 GDIP01094273 GDIP01071324 JAM32391.1 GDIQ01143613 JAL08113.1 GDIQ01045852 JAN48885.1 GDIQ01137908 JAL13818.1 GDIP01109072 JAL94642.1 GDIQ01094524 JAL57202.1 GDIQ01124964 JAL26762.1 GDIP01109073 JAL94641.1 GDIP01060827 JAM42888.1 GDIP01155164 JAJ68238.1 GDIQ01213232 JAK38493.1 GDIQ01140109 JAL11617.1 GDIP01127315 JAL76399.1 GDIQ01107819 JAL43907.1 UYYA01004098 VDM59544.1 GDIP01190968 JAJ32434.1 GDIQ01107818 JAL43908.1 GDIQ01210932 JAK40793.1 JI164624 ADY40437.1 GDIQ01085533 JAN09204.1 DS268446 EFP02182.1 GDIQ01094523 JAL57203.1 NIPN01000136 OZG09712.1 GDIP01143565 JAJ79837.1
KQ414786 KOC60813.1 KQ434896 KZC10742.1 KZ288391 PBC26357.1 ADTU01024457 ADTU01024458 KQ981208 KYN44656.1 KB632100 ERL88846.1 APGK01041757 KB740997 ENN75845.1 GL449658 EFN82290.1 QOIP01000010 RLU17593.1 KK107374 EZA51596.1 UFQT01001761 SSX31693.1 SSX31692.1 UFQT01002921 SSX34289.1 KQ435789 KOX74487.1 GDIP01179215 JAJ44187.1 GDIP01194666 JAJ28736.1 GDIP01201817 GDIP01111418 GDIP01044134 JAL92296.1 GDIP01199161 JAJ24241.1 GDIP01072629 JAM31086.1 GDIP01122822 JAL80892.1 GDIP01084523 JAM19192.1 GDIQ01007066 JAN87671.1 GDIP01205511 JAJ17891.1 GDIP01199162 JAJ24240.1 GDIP01203361 JAJ20041.1 GDIP01235107 JAI88294.1 GDIP01046881 JAM56834.1 GDIQ01069980 JAN24757.1 GDIP01084525 JAM19190.1 GDIP01012168 JAM91547.1 GDIP01084524 JAM19191.1 LRGB01002849 KZS06140.1 GDIP01124820 JAL78894.1 GDIP01048578 JAM55137.1 AMQN01005970 KB297235 ELU10759.1 GDIP01251954 JAI71447.1 GDIQ01224532 JAK27193.1 KB201802 ESO94506.1 GDIP01086026 JAM17689.1 GDIP01084526 JAM19189.1 GDIP01130323 JAL73391.1 GDIQ01203334 JAK48391.1 GDIP01094273 GDIP01071324 JAM32391.1 GDIQ01143613 JAL08113.1 GDIQ01045852 JAN48885.1 GDIQ01137908 JAL13818.1 GDIP01109072 JAL94642.1 GDIQ01094524 JAL57202.1 GDIQ01124964 JAL26762.1 GDIP01109073 JAL94641.1 GDIP01060827 JAM42888.1 GDIP01155164 JAJ68238.1 GDIQ01213232 JAK38493.1 GDIQ01140109 JAL11617.1 GDIP01127315 JAL76399.1 GDIQ01107819 JAL43907.1 UYYA01004098 VDM59544.1 GDIP01190968 JAJ32434.1 GDIQ01107818 JAL43908.1 GDIQ01210932 JAK40793.1 JI164624 ADY40437.1 GDIQ01085533 JAN09204.1 DS268446 EFP02182.1 GDIQ01094523 JAL57203.1 NIPN01000136 OZG09712.1 GDIP01143565 JAJ79837.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2H1WHD1
A0A194PXK2
A0A1W5YN02
A0A0L7QQA0
A0A088ALS8
A0A154PFZ0
+ More
A0A2A3E3Q0 A0A158NS37 A0A195FW52 U4UE49 N6U2Q0 E2BPT9 A0A3L8DBU5 A0A026W7M0 A0A336MSF1 A0A336MMQ1 A0A336MYR3 A0A0M9A074 A0A0P5BVX3 A0A0P5BV94 A0A0P5ULF6 A0A0N7ZYU7 A0A0P5XGQ3 A0A0P5TY23 A0A0P5WNZ0 A0A0P6JIF6 A0A0P5A0R2 A0A0P5AGJ5 A0A0N7ZXC2 A0A0P4XYG0 A0A0P5ZAJ1 A0A0N8E1W1 A0A0P5XH45 A0A0P6C7D8 A0A0P5WLK6 A0A164NPU1 A0A0P5TL70 A0A0P5Z581 R7UWR3 A0A0P4X136 A0A0P5HQU5 V4ALP1 A0A0P5WIG7 A0A0P5WLX8 A0A3Q3XM66 A0A0P5TKV9 A0A0P5JL96 A0A0P5XPT4 A0A0P5NL68 A0A0N8EAH7 A0A0P5NYQ2 A0A0P5UWS3 A0A0P5SET8 A0A0P5QE56 A0A0P5UV70 A0A0P5Z6R4 A0A0P5DYV0 A0A0P5INE5 A0A0P5NN30 A0A0P5TT68 A0A0N8C7A5 A0A0R3PR76 A0A0P5B2G2 A0A0P5R4J9 A0A0N5AQW6 A0A0P5J4V2 F1KRD3 A0A0P6CYY7 E3MHZ4 A0A0N8CC17 A0A261BHA1 A0A0P5EPF5
A0A2A3E3Q0 A0A158NS37 A0A195FW52 U4UE49 N6U2Q0 E2BPT9 A0A3L8DBU5 A0A026W7M0 A0A336MSF1 A0A336MMQ1 A0A336MYR3 A0A0M9A074 A0A0P5BVX3 A0A0P5BV94 A0A0P5ULF6 A0A0N7ZYU7 A0A0P5XGQ3 A0A0P5TY23 A0A0P5WNZ0 A0A0P6JIF6 A0A0P5A0R2 A0A0P5AGJ5 A0A0N7ZXC2 A0A0P4XYG0 A0A0P5ZAJ1 A0A0N8E1W1 A0A0P5XH45 A0A0P6C7D8 A0A0P5WLK6 A0A164NPU1 A0A0P5TL70 A0A0P5Z581 R7UWR3 A0A0P4X136 A0A0P5HQU5 V4ALP1 A0A0P5WIG7 A0A0P5WLX8 A0A3Q3XM66 A0A0P5TKV9 A0A0P5JL96 A0A0P5XPT4 A0A0P5NL68 A0A0N8EAH7 A0A0P5NYQ2 A0A0P5UWS3 A0A0P5SET8 A0A0P5QE56 A0A0P5UV70 A0A0P5Z6R4 A0A0P5DYV0 A0A0P5INE5 A0A0P5NN30 A0A0P5TT68 A0A0N8C7A5 A0A0R3PR76 A0A0P5B2G2 A0A0P5R4J9 A0A0N5AQW6 A0A0P5J4V2 F1KRD3 A0A0P6CYY7 E3MHZ4 A0A0N8CC17 A0A261BHA1 A0A0P5EPF5
PDB
5D06
E-value=1.07926e-104,
Score=977
Ontologies
PATHWAY
GO
PANTHER
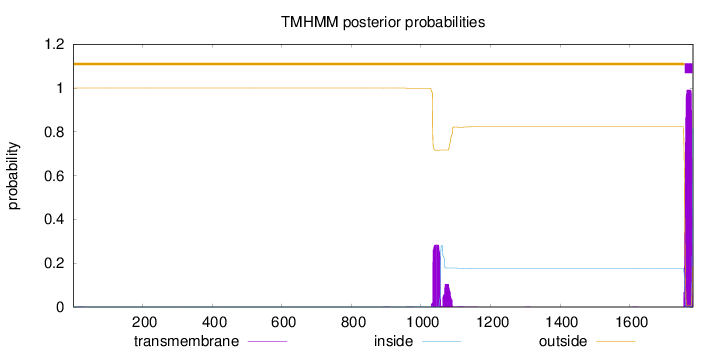
Topology
Length:
1782
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
30.31264
Exp number, first 60 AAs:
0.00344
Total prob of N-in:
0.00029
outside
1 - 1758
TMhelix
1759 - 1781
inside
1782 - 1782
Population Genetic Test Statistics
Pi
21.264481
Theta
20.726809
Tajima's D
-1.032945
CLR
0.952343
CSRT
0.133643317834108
Interpretation
Uncertain
