Gene
KWMTBOMO13451
Pre Gene Modal
BGIBMGA010865
Annotation
PREDICTED:_probable_cytochrome_P450_313a1?_partial_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.124 Mitochondrial Reliability : 1.636
Sequence
CDS
ATGATCATCGCCGGCACCGATACTTCGGCCATCGGTATATCCTTCACGTTACTCATGCTGGCCCGATATCCTGATGTACAAGAGAAAGTCTACCAGGAGATCCAGGAATTGTTCGGTGACTCAGAGAGGCCGCCCGAAGTGGAAGACATCCATCGTCTTCTGTACTTGGATGCAGTGATCAAAGAAGCGATGCGATTATATCCTCCGGCGCCAGTCATCATAAGGAAAGTCGAAAGAGAAACTAAACTGCCATCCGGATTAATACTGCCGCGGGACATTGGGATTTTAATACCAATCTGGTCCGTGAACCGCAATCCTAAGTACTGGGGCGACGACGCCGATGTATTTAGACCCGAGCGGTTTTTGGATGGTACCAAAAAACATCCAACAGCATTCATGACGTTTAGCCAGGGACCGAGAGCTTGTCTAGGCTTTAAGTTCGCTATGAACTCAGCTAAGGCGGCTCTGGCTTCCATCCTTCGCCATTACCGCATCAAACCACCGTCAGAACTGACCTCAATGTCCCCAGGACAATACCCTCCCATCAGGGTCAAGTTTGCCCTAATGACGAGGGACGTTGATAATTTCAGAATACAATTAGAATCTAGATCGTAA
Protein
MIIAGTDTSAIGISFTLLMLARYPDVQEKVYQEIQELFGDSERPPEVEDIHRLLYLDAVIKEAMRLYPPAPVIIRKVERETKLPSGLILPRDIGILIPIWSVNRNPKYWGDDADVFRPERFLDGTKKHPTAFMTFSQGPRACLGFKFAMNSAKAALASILRHYRIKPPSELTSMSPGQYPPIRVKFALMTRDVDNFRIQLESRS
Summary
Cofactor
heme
Similarity
Belongs to the cytochrome P450 family.
Uniprot
H9J588
L0N785
A0A2A4JGE5
A0A2H1VMK4
A0A286QUH1
A0A0D6A1C6
+ More
A0A2A4JHV7 A0A2A4JGQ9 A0A068EVJ0 A0A2Z5SAV1 A0A248QED4 A0A0L7KP65 A0A068EU38 A0A0K8TUI4 A0A0L7KSI3 A0A0K8TUL8 A0A0K8TV26 A0A2H1WUL9 A0A2A4IT41 A0A2H1VP75 A0A3S2PCK4 A0A1E1WU74 A0A0C5C579 H9J5D0 A0A0K8TVP5 A0A0K8TV31 A0A194PYM9 H9J5C7 A0A212FEU5 H9J5C8 A0A194Q3L1 A0A0L7L4L4 A0A194R0G6 A0A0K8TUH6 A0A2H1WBM8 A0A1V0D9H7 A0A194REI2 A0A2A4ISM9 A0A212EGV4 X5DB16 A0A194PZG6 X5DB21 A0A194PY94 A0A194R208 A0A194R193 A0A2H1WUI5 A0A3S2TR28 A0A2H1WBK5 S4VGT4 A0A2A4IYA1 A0A248QEI3 A0A194RF00 Q4R1I7 J7FIT6 A0A346IHZ3 A0A1V0D9H9 A0A346II01 A0A194R5Z5 A0A0L7KML1 A0A3S2M7I8 H9J5D1 A0A1V0D9I5 A0A146KWM2 H9J5C6 A0A0L7KP31 A0A0U2DV61 A0A2U7QTF3 A0A2W1BZ42 A0A2A4JY96 A0A194PZ23 A0A0K8SV38 A0A0K8SV22 A0A2H1V533 A0A0K0D2Y2 A0A146MAJ2 A0A068EU45 A0A158PF81 A0A0C5C571 A0A016U9I0 H9JHK8 L0N5K2 L7STW5 A0A212EQ79 C1IDD0 A0A158QQ30 A0A194QLZ7 A0A0B2VPC8 A0A0N1IEQ4 A0A182S8E5 A0A194QA50 A0A0P8ZXK0 A0A0K8TVT3 A0A182M2E4 A0A2A4JMP1 A0A182GC09
A0A2A4JHV7 A0A2A4JGQ9 A0A068EVJ0 A0A2Z5SAV1 A0A248QED4 A0A0L7KP65 A0A068EU38 A0A0K8TUI4 A0A0L7KSI3 A0A0K8TUL8 A0A0K8TV26 A0A2H1WUL9 A0A2A4IT41 A0A2H1VP75 A0A3S2PCK4 A0A1E1WU74 A0A0C5C579 H9J5D0 A0A0K8TVP5 A0A0K8TV31 A0A194PYM9 H9J5C7 A0A212FEU5 H9J5C8 A0A194Q3L1 A0A0L7L4L4 A0A194R0G6 A0A0K8TUH6 A0A2H1WBM8 A0A1V0D9H7 A0A194REI2 A0A2A4ISM9 A0A212EGV4 X5DB16 A0A194PZG6 X5DB21 A0A194PY94 A0A194R208 A0A194R193 A0A2H1WUI5 A0A3S2TR28 A0A2H1WBK5 S4VGT4 A0A2A4IYA1 A0A248QEI3 A0A194RF00 Q4R1I7 J7FIT6 A0A346IHZ3 A0A1V0D9H9 A0A346II01 A0A194R5Z5 A0A0L7KML1 A0A3S2M7I8 H9J5D1 A0A1V0D9I5 A0A146KWM2 H9J5C6 A0A0L7KP31 A0A0U2DV61 A0A2U7QTF3 A0A2W1BZ42 A0A2A4JY96 A0A194PZ23 A0A0K8SV38 A0A0K8SV22 A0A2H1V533 A0A0K0D2Y2 A0A146MAJ2 A0A068EU45 A0A158PF81 A0A0C5C571 A0A016U9I0 H9JHK8 L0N5K2 L7STW5 A0A212EQ79 C1IDD0 A0A158QQ30 A0A194QLZ7 A0A0B2VPC8 A0A0N1IEQ4 A0A182S8E5 A0A194QA50 A0A0P8ZXK0 A0A0K8TVT3 A0A182M2E4 A0A2A4JMP1 A0A182GC09
Pubmed
EMBL
BABH01039691
AB436841
BAM73795.1
NWSH01001465
PCG71165.1
ODYU01003376
+ More
SOQ42053.1 KX443471 ASO98046.1 AB795798 BAQ56325.1 PCG71164.1 PCG71161.1 KM016716 AID54868.1 LC326250 BBA58211.1 KX425016 ASN63843.1 JTDY01007865 KOB64900.1 KM016717 AID54869.1 GCVX01000088 JAI18142.1 JTDY01006414 KOB66031.1 GCVX01000053 JAI18177.1 GCVX01000092 JAI18138.1 ODYU01011123 SOQ56666.1 NWSH01007729 PCG62839.1 SOQ42054.1 RSAL01000100 RVE47555.1 GDQN01000461 JAT90593.1 KP001145 AJN91190.1 BABH01037064 BABH01037065 GCVX01000090 JAI18140.1 GCVX01000087 JAI18143.1 KQ459586 KPI98133.1 BABH01037061 AGBW02008898 OWR52238.1 KPI98000.1 JTDY01002945 KOB70438.1 KQ460883 KPJ11283.1 GCVX01000086 JAI18144.1 ODYU01007567 SOQ50447.1 KY212088 ARA91652.1 KQ460323 KPJ15874.1 NWSH01008140 PCG62679.1 AGBW02015005 OWR40717.1 KF701170 AHW57340.1 KPI98134.1 KF701175 AHW57345.1 KPI97998.1 KPJ11285.1 KPJ11284.1 SOQ56667.1 RSAL01000018 RVE52820.1 SOQ50448.1 KC789756 AGO62011.1 NWSH01005164 PCG64388.1 KX443470 ASO98045.1 KPJ15875.1 AB201381 BAD99563.1 JX310094 AFP20605.1 MH138207 AXP17148.1 KY212090 ARA91654.1 MH138215 AXP17156.1 KPJ11286.1 JTDY01009042 KOB64169.1 RVE52821.1 BABH01037067 BABH01037068 KY212089 ARA91653.1 GDHC01018604 JAQ00025.1 BABH01037058 BABH01037059 JTDY01007639 KOB65052.1 KP091899 AKQ00287.1 KY348795 AUX13052.1 KZ149894 PZC78854.1 NWSH01000444 PCG76362.1 KPI97999.1 GBRD01008662 GBRD01008659 JAG57162.1 GBRD01008658 JAG57163.1 ODYU01000688 SOQ35886.1 GDHC01006464 GDHC01006386 GDHC01002833 JAQ12165.1 JAQ12243.1 JAQ15796.1 KM016722 AID54874.1 UYYA01000750 VDM54593.1 KP001140 AJN91185.1 JARK01001387 EYC11273.1 BABH01012758 AK343188 BAM73878.1 JN160688 AGC22877.1 AGBW02013301 OWR43639.1 EU546147 ACD76734.1 UZAF01018368 VDO50926.1 KQ458981 KPJ04476.1 JPKZ01001207 UYWY01023609 KHN83257.1 VDM47778.1 KQ460882 KPJ11479.1 KQ459299 KPJ01870.1 CH902617 KPU79450.1 GCVX01000055 JAI18175.1 AXCM01008551 NWSH01001018 PCG73089.1 JXUM01053525 KQ561780 KXJ77562.1
SOQ42053.1 KX443471 ASO98046.1 AB795798 BAQ56325.1 PCG71164.1 PCG71161.1 KM016716 AID54868.1 LC326250 BBA58211.1 KX425016 ASN63843.1 JTDY01007865 KOB64900.1 KM016717 AID54869.1 GCVX01000088 JAI18142.1 JTDY01006414 KOB66031.1 GCVX01000053 JAI18177.1 GCVX01000092 JAI18138.1 ODYU01011123 SOQ56666.1 NWSH01007729 PCG62839.1 SOQ42054.1 RSAL01000100 RVE47555.1 GDQN01000461 JAT90593.1 KP001145 AJN91190.1 BABH01037064 BABH01037065 GCVX01000090 JAI18140.1 GCVX01000087 JAI18143.1 KQ459586 KPI98133.1 BABH01037061 AGBW02008898 OWR52238.1 KPI98000.1 JTDY01002945 KOB70438.1 KQ460883 KPJ11283.1 GCVX01000086 JAI18144.1 ODYU01007567 SOQ50447.1 KY212088 ARA91652.1 KQ460323 KPJ15874.1 NWSH01008140 PCG62679.1 AGBW02015005 OWR40717.1 KF701170 AHW57340.1 KPI98134.1 KF701175 AHW57345.1 KPI97998.1 KPJ11285.1 KPJ11284.1 SOQ56667.1 RSAL01000018 RVE52820.1 SOQ50448.1 KC789756 AGO62011.1 NWSH01005164 PCG64388.1 KX443470 ASO98045.1 KPJ15875.1 AB201381 BAD99563.1 JX310094 AFP20605.1 MH138207 AXP17148.1 KY212090 ARA91654.1 MH138215 AXP17156.1 KPJ11286.1 JTDY01009042 KOB64169.1 RVE52821.1 BABH01037067 BABH01037068 KY212089 ARA91653.1 GDHC01018604 JAQ00025.1 BABH01037058 BABH01037059 JTDY01007639 KOB65052.1 KP091899 AKQ00287.1 KY348795 AUX13052.1 KZ149894 PZC78854.1 NWSH01000444 PCG76362.1 KPI97999.1 GBRD01008662 GBRD01008659 JAG57162.1 GBRD01008658 JAG57163.1 ODYU01000688 SOQ35886.1 GDHC01006464 GDHC01006386 GDHC01002833 JAQ12165.1 JAQ12243.1 JAQ15796.1 KM016722 AID54874.1 UYYA01000750 VDM54593.1 KP001140 AJN91185.1 JARK01001387 EYC11273.1 BABH01012758 AK343188 BAM73878.1 JN160688 AGC22877.1 AGBW02013301 OWR43639.1 EU546147 ACD76734.1 UZAF01018368 VDO50926.1 KQ458981 KPJ04476.1 JPKZ01001207 UYWY01023609 KHN83257.1 VDM47778.1 KQ460882 KPJ11479.1 KQ459299 KPJ01870.1 CH902617 KPU79450.1 GCVX01000055 JAI18175.1 AXCM01008551 NWSH01001018 PCG73089.1 JXUM01053525 KQ561780 KXJ77562.1
Proteomes
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
H9J588
L0N785
A0A2A4JGE5
A0A2H1VMK4
A0A286QUH1
A0A0D6A1C6
+ More
A0A2A4JHV7 A0A2A4JGQ9 A0A068EVJ0 A0A2Z5SAV1 A0A248QED4 A0A0L7KP65 A0A068EU38 A0A0K8TUI4 A0A0L7KSI3 A0A0K8TUL8 A0A0K8TV26 A0A2H1WUL9 A0A2A4IT41 A0A2H1VP75 A0A3S2PCK4 A0A1E1WU74 A0A0C5C579 H9J5D0 A0A0K8TVP5 A0A0K8TV31 A0A194PYM9 H9J5C7 A0A212FEU5 H9J5C8 A0A194Q3L1 A0A0L7L4L4 A0A194R0G6 A0A0K8TUH6 A0A2H1WBM8 A0A1V0D9H7 A0A194REI2 A0A2A4ISM9 A0A212EGV4 X5DB16 A0A194PZG6 X5DB21 A0A194PY94 A0A194R208 A0A194R193 A0A2H1WUI5 A0A3S2TR28 A0A2H1WBK5 S4VGT4 A0A2A4IYA1 A0A248QEI3 A0A194RF00 Q4R1I7 J7FIT6 A0A346IHZ3 A0A1V0D9H9 A0A346II01 A0A194R5Z5 A0A0L7KML1 A0A3S2M7I8 H9J5D1 A0A1V0D9I5 A0A146KWM2 H9J5C6 A0A0L7KP31 A0A0U2DV61 A0A2U7QTF3 A0A2W1BZ42 A0A2A4JY96 A0A194PZ23 A0A0K8SV38 A0A0K8SV22 A0A2H1V533 A0A0K0D2Y2 A0A146MAJ2 A0A068EU45 A0A158PF81 A0A0C5C571 A0A016U9I0 H9JHK8 L0N5K2 L7STW5 A0A212EQ79 C1IDD0 A0A158QQ30 A0A194QLZ7 A0A0B2VPC8 A0A0N1IEQ4 A0A182S8E5 A0A194QA50 A0A0P8ZXK0 A0A0K8TVT3 A0A182M2E4 A0A2A4JMP1 A0A182GC09
A0A2A4JHV7 A0A2A4JGQ9 A0A068EVJ0 A0A2Z5SAV1 A0A248QED4 A0A0L7KP65 A0A068EU38 A0A0K8TUI4 A0A0L7KSI3 A0A0K8TUL8 A0A0K8TV26 A0A2H1WUL9 A0A2A4IT41 A0A2H1VP75 A0A3S2PCK4 A0A1E1WU74 A0A0C5C579 H9J5D0 A0A0K8TVP5 A0A0K8TV31 A0A194PYM9 H9J5C7 A0A212FEU5 H9J5C8 A0A194Q3L1 A0A0L7L4L4 A0A194R0G6 A0A0K8TUH6 A0A2H1WBM8 A0A1V0D9H7 A0A194REI2 A0A2A4ISM9 A0A212EGV4 X5DB16 A0A194PZG6 X5DB21 A0A194PY94 A0A194R208 A0A194R193 A0A2H1WUI5 A0A3S2TR28 A0A2H1WBK5 S4VGT4 A0A2A4IYA1 A0A248QEI3 A0A194RF00 Q4R1I7 J7FIT6 A0A346IHZ3 A0A1V0D9H9 A0A346II01 A0A194R5Z5 A0A0L7KML1 A0A3S2M7I8 H9J5D1 A0A1V0D9I5 A0A146KWM2 H9J5C6 A0A0L7KP31 A0A0U2DV61 A0A2U7QTF3 A0A2W1BZ42 A0A2A4JY96 A0A194PZ23 A0A0K8SV38 A0A0K8SV22 A0A2H1V533 A0A0K0D2Y2 A0A146MAJ2 A0A068EU45 A0A158PF81 A0A0C5C571 A0A016U9I0 H9JHK8 L0N5K2 L7STW5 A0A212EQ79 C1IDD0 A0A158QQ30 A0A194QLZ7 A0A0B2VPC8 A0A0N1IEQ4 A0A182S8E5 A0A194QA50 A0A0P8ZXK0 A0A0K8TVT3 A0A182M2E4 A0A2A4JMP1 A0A182GC09
PDB
6C93
E-value=3.0542e-25,
Score=282
Ontologies
GO
Topology
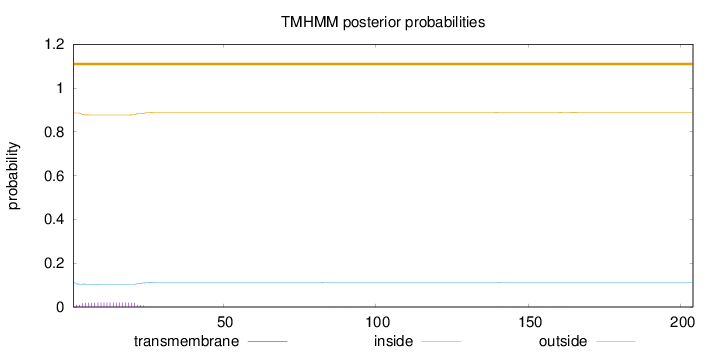
Length:
204
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.43963
Exp number, first 60 AAs:
0.40168
Total prob of N-in:
0.11454
outside
1 - 204
Population Genetic Test Statistics
Pi
15.593858
Theta
17.249848
Tajima's D
-0.382763
CLR
10.17066
CSRT
0.272186390680466
Interpretation
Uncertain
