Gene
KWMTBOMO13448
Pre Gene Modal
BGIBMGA010864
Annotation
Aspartyl-tRNA_synthetase?_mitochondrial_[Papilio_machaon]
Full name
Aspartate--tRNA ligase
+ More
Aspartate--tRNA(Asp/Asn) ligase
Aspartate--tRNA(Asp) ligase
Aspartate--tRNA(Asp/Asn) ligase
Aspartate--tRNA(Asp) ligase
Alternative Name
Aspartyl-tRNA synthetase
Non-discriminating aspartyl-tRNA synthetase
Discriminating aspartyl-tRNA synthetase
Aspartyl-tRNA synthetase 1
Non-discriminating aspartyl-tRNA synthetase
Discriminating aspartyl-tRNA synthetase
Aspartyl-tRNA synthetase 1
Location in the cell
Cytoplasmic Reliability : 1.968 Nuclear Reliability : 1.864
Sequence
CDS
ATGGCTGGATAGACTCACACATGCGGCGAACTGAGACCTAAGAATGTCGGTGAAAGAGTCGTCATATGCGGCTGGGTGCAGTATTCACGTCTGTCCAAATTTTTACTGCTTCGAGACGCCTACGGACTCGCACAATGTATTGTTGGTTGTGACAATATTGATTTATCCAGCTTACAACTAGAATCTGTAGTACAAATTGAAGGCATGGTTTCCATAAGGCCCGGAGACACAGTGAACCCTGAAATGGCTACCGGTGAAATTGAGAAACCAAAAGAACAATTGAGAATGCAACACAGATACATTGATTTAAGATTTCCCGTGATGCAAAATAATTTAAGAACTAGATCACAGATGCTGCATAAGATGCGGAGATTTCTAGTGGAAAATTATGGGTTTATCGAAGTTGAGACTCCAACACTTTTCTGCAGGACACCAGGTGGTGCTAGAGAATTTGTTGTCCCCACTCGTCATGAAGGATTGTTCTATTCTTTAGTCCAAAGTCCTCAACAATTCAAACAAATGCTTATGTCTGGTGGAATAGACCGGTATTTCCAAGTAGCCAGGTGCTACAGAGCTACCACTCGTCCAGACCGTCAACCAGAATTTACTCAGTTGGATATTGAACTATCTTTTACCACTTTAGAAGGAGTTCTAACATTGATTGAAGAGATGCTATATGATACTTTTACGAAACCATTACCAAGACCTCCATTTAATAGAATTACATATAAAGAAGCACTGGAAAATTATGGAAGTGATAAACCAAATTTACTGTATGATTTGAAATTTATAAACATCAAACATTTATTTGACAAAAAAGAAGATGACAACTTTGGTGCTCTCCTGCTACCTTACCCCAGTGAGATGGGGAAGCTTACCACCAAATACAAAGAGAAAGTAAATGAAATACAAGAGAGAATACAAGATGCCATTGGTCAGAATCGTGCTGCAATTATATCTATAAGCGAAGACACTGAAAATGCCTGCCTATGTTCAGGAGAAATTAAAGAAATAATAATATCAGTATTGAAATCTAAAGAGCTGCTAAAAGTTAATGACAATGTTGCGCCTCTGTGGCTGATCGACTTCCCGTTATTTGTGAAAGGAGATGATGGACTTCAACCATGCCATCATCCCTTTACAGCACCTCATCCGGATGACCTACATTTATTAGAAACTGATCCATTGAAAGTTCGGTCATTAGCATATGATTTAGTGATGAATGGTAATGAAATAGGAGGTGGTTCAGTCCGAATACACAGTCCGCATTTACAAGTTAAAATTATGAACATGTTGAACATAGATCCAGAGAAATTGAGGCATTTTGTGAATGCATTAAGTAGTGGCTGTCCTCCTCATGCCGGCATTGCTCTTGGTATTGACAGGTTAATAGCAATTGCCTGTGATGCAGAGTCAATAAGAGACGTGATTGCATTCCCCAAGTCTCATGATGGCCGTGATCCTCTGTCAGGTGCACCTAATGTTTTATCACATAGTGATAGAAATTATTATCATTTATATCATATTAAATGA
Protein
MAGXTHTCGELRPKNVGERVVICGWVQYSRLSKFLLLRDAYGLAQCIVGCDNIDLSSLQLESVVQIEGMVSIRPGDTVNPEMATGEIEKPKEQLRMQHRYIDLRFPVMQNNLRTRSQMLHKMRRFLVENYGFIEVETPTLFCRTPGGAREFVVPTRHEGLFYSLVQSPQQFKQMLMSGGIDRYFQVARCYRATTRPDRQPEFTQLDIELSFTTLEGVLTLIEEMLYDTFTKPLPRPPFNRITYKEALENYGSDKPNLLYDLKFINIKHLFDKKEDDNFGALLLPYPSEMGKLTTKYKEKVNEIQERIQDAIGQNRAAIISISEDTENACLCSGEIKEIIISVLKSKELLKVNDNVAPLWLIDFPLFVKGDDGLQPCHHPFTAPHPDDLHLLETDPLKVRSLAYDLVMNGNEIGGGSVRIHSPHLQVKIMNMLNIDPEKLRHFVNALSSGCPPHAGIALGIDRLIAIACDAESIRDVIAFPKSHDGRDPLSGAPNVLSHSDRNYYHLYHIK
Summary
Description
Catalyzes the attachment of L-aspartate to tRNA(Asp) in a two-step reaction: L-aspartate is first activated by ATP to form Asp-AMP and then transferred to the acceptor end of tRNA(Asp).
Aspartyl-tRNA synthetase with relaxed tRNA specificity since it is able to aspartylate not only its cognate tRNA(Asp) but also tRNA(Asn). Reaction proceeds in two steps: L-aspartate is first activated by ATP to form Asp-AMP and then transferred to the acceptor end of tRNA(Asp/Asn).
Catalyzes the attachment of L-aspartate to tRNA(Asp) in a two-step reaction: L-aspartate is first activated by ATP to form Asp-AMP and then transferred to the acceptor end of tRNA(Asp). Is specific for tRNA(Asp) since it aspartylates tRNA(Asn) 3 orders of magnitude less efficiently than tRNA(Asp).
Aspartyl-tRNA synthetase with relaxed tRNA specificity since it is able to aspartylate not only its cognate tRNA(Asp) but also tRNA(Asn). Reaction proceeds in two steps: L-aspartate is first activated by ATP to form Asp-AMP and then transferred to the acceptor end of tRNA(Asp/Asn).
Catalyzes the attachment of L-aspartate to tRNA(Asp) in a two-step reaction: L-aspartate is first activated by ATP to form Asp-AMP and then transferred to the acceptor end of tRNA(Asp). Is specific for tRNA(Asp) since it aspartylates tRNA(Asn) 3 orders of magnitude less efficiently than tRNA(Asp).
Catalytic Activity
ATP + L-aspartate + tRNA(Asp) = AMP + diphosphate + L-aspartyl-tRNA(Asp)
ATP + L-aspartate + tRNA(Asx) = AMP + diphosphate + L-aspartyl-tRNA(Asx)
ATP + L-aspartate + tRNA(Asx) = AMP + diphosphate + L-aspartyl-tRNA(Asx)
Biophysicochemical Properties
9 uM for L-aspartate (at 37 degrees Celsius)
30 uM for L-aspartate (at 70 degrees Celsius)
120 uM for ATP (at 37 degrees Celsius)
280 uM for ATP (at 70 degrees Celsius)
0.044 uM for tRNA(Asp) (at 37 degrees Celsius)
0.030 uM for tRNA(Asp) (at 70 degrees Celsius)
3.4 uM for tRNA(Asn) (at 70 degrees Celsius)
30 uM for L-aspartate (at 70 degrees Celsius)
120 uM for ATP (at 37 degrees Celsius)
280 uM for ATP (at 70 degrees Celsius)
0.044 uM for tRNA(Asp) (at 37 degrees Celsius)
0.030 uM for tRNA(Asp) (at 70 degrees Celsius)
3.4 uM for tRNA(Asn) (at 70 degrees Celsius)
Subunit
Homodimer.
Similarity
Belongs to the class-II aminoacyl-tRNA synthetase family. Type 1 subfamily.
Keywords
Aminoacyl-tRNA synthetase
ATP-binding
Complete proteome
Cytoplasm
Ligase
Nucleotide-binding
Protein biosynthesis
Reference proteome
3D-structure
Direct protein sequencing
Feature
chain Aspartate--tRNA(Asp/Asn) ligase
Uniprot
H9JMW1
A0A2W1BQF6
A0A2A4JII1
H9JMW0
A0A194R149
A0A0L7KUU8
+ More
A0A212EIN8 A0A194PXJ4 A0A2H1X3S0 D6WJB0 A0A0C9QZB7 A0A0C9QGP4 A0A1W4WKH8 A0A0V0GC92 A0A0A1WV06 A0A0A1X7W5 E9J836 A0A1B6C2E3 A0A1B6CB86 B4P990 B3NNL7 Q9VJH2 A0A151ITX3 A0A0Q9WAC6 B4MYY1 A0A3L8DGI4 A0A0J9R3K6 B4I559 B4LQX8 B4JQX0 E2A1R8 B3MK45 B4KJR2 A0A2P8XTY3 A0A0M4ERI2 E0VEP3 A0A069DVL5 A0A154PNX3 A0A182L2D1 A0A182IEL3 A0A182TP04 A0A182VIH7 G1KR37 A0A3B3YKL2 R4G9Q0 A0A3B3VMQ4 A0A3B4B1L7 A0A2K5R7R8 A0A0L7RG52 A0A2S2ND19 A0A0N0U498 A0A182QZW8 F1S742 A0A286ZWZ5 H1A3Z9 A0A3B3IT01 A0A091UX00 A0A2K5R7R5 J9K6D5 H2SYM8 A0A091NXS1 A0A091LQU4 A0A087ZV42 A0A2A3E8F3 S4RXF5 A0A3Q3XHH8 A0A3Q0IWP3 E2C360 A0A182WTH7 A0A182NLM4 A0A3B3RWS5 A0A3Q3XMC5 A0A3B3IS01 A0A3Q7PIA3 A0A2I0MBS3 A0A2H0D9F6 A0A093FBC9 A0A218V4N5 A0A2D4FF60 A0A0A1NXR9 A0A0R3TVN0 A0A367KUF4 A0A3P4AQ79 A0A0G2Z583 B5YDT0 A0A2E0RP97 A0A0M3I244 A0A0J7L1N1 A0A0N4ZSP0 L9KIG0 A0A3M1QHE6 A0A3M1LUE7 Q2BI63 A0A2M8PTS7 A0A2E5XYG8 A0A1J1EN87 B8E1C0 P36419 Q5SKD2 Q72KH6
A0A212EIN8 A0A194PXJ4 A0A2H1X3S0 D6WJB0 A0A0C9QZB7 A0A0C9QGP4 A0A1W4WKH8 A0A0V0GC92 A0A0A1WV06 A0A0A1X7W5 E9J836 A0A1B6C2E3 A0A1B6CB86 B4P990 B3NNL7 Q9VJH2 A0A151ITX3 A0A0Q9WAC6 B4MYY1 A0A3L8DGI4 A0A0J9R3K6 B4I559 B4LQX8 B4JQX0 E2A1R8 B3MK45 B4KJR2 A0A2P8XTY3 A0A0M4ERI2 E0VEP3 A0A069DVL5 A0A154PNX3 A0A182L2D1 A0A182IEL3 A0A182TP04 A0A182VIH7 G1KR37 A0A3B3YKL2 R4G9Q0 A0A3B3VMQ4 A0A3B4B1L7 A0A2K5R7R8 A0A0L7RG52 A0A2S2ND19 A0A0N0U498 A0A182QZW8 F1S742 A0A286ZWZ5 H1A3Z9 A0A3B3IT01 A0A091UX00 A0A2K5R7R5 J9K6D5 H2SYM8 A0A091NXS1 A0A091LQU4 A0A087ZV42 A0A2A3E8F3 S4RXF5 A0A3Q3XHH8 A0A3Q0IWP3 E2C360 A0A182WTH7 A0A182NLM4 A0A3B3RWS5 A0A3Q3XMC5 A0A3B3IS01 A0A3Q7PIA3 A0A2I0MBS3 A0A2H0D9F6 A0A093FBC9 A0A218V4N5 A0A2D4FF60 A0A0A1NXR9 A0A0R3TVN0 A0A367KUF4 A0A3P4AQ79 A0A0G2Z583 B5YDT0 A0A2E0RP97 A0A0M3I244 A0A0J7L1N1 A0A0N4ZSP0 L9KIG0 A0A3M1QHE6 A0A3M1LUE7 Q2BI63 A0A2M8PTS7 A0A2E5XYG8 A0A1J1EN87 B8E1C0 P36419 Q5SKD2 Q72KH6
EC Number
6.1.1.12
6.1.1.23
6.1.1.23
Pubmed
19121390
28756777
26354079
26227816
22118469
18362917
+ More
19820115 25830018 21282665 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 22936249 18057021 20798317 29403074 20566863 26334808 20966253 25463417 20360741 16710414 19608861 21269460 23186163 24275569 25944712 21551351 29240929 23371554 29674435 29337314 23385571 28066333 8319804 8045252 7966328 10843857 11914489 9220965 10727213 15064768
19820115 25830018 21282665 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 22936249 18057021 20798317 29403074 20566863 26334808 20966253 25463417 20360741 16710414 19608861 21269460 23186163 24275569 25944712 21551351 29240929 23371554 29674435 29337314 23385571 28066333 8319804 8045252 7966328 10843857 11914489 9220965 10727213 15064768
EMBL
BABH01029070
BABH01029071
KZ149981
PZC75805.1
NWSH01001352
PCG71576.1
+ More
BABH01029075 BABH01029076 KQ460883 KPJ11239.1 JTDY01005408 KOB67042.1 AGBW02014600 OWR41357.1 KQ459586 KPI98047.1 ODYU01013276 SOQ59983.1 KQ971343 EFA03147.2 GBYB01006062 JAG75829.1 GBYB01013753 JAG83520.1 GECL01001117 JAP05007.1 GBXI01011403 JAD02889.1 GBXI01007502 JAD06790.1 GL768788 EFZ11007.1 GEDC01029607 JAS07691.1 GEDC01026550 JAS10748.1 CM000158 EDW90219.2 CH954179 EDV56668.2 AE014134 AY119180 AAF53577.3 AAM51040.1 AGB93054.1 AGB93055.1 KQ981004 KYN10393.1 CH940649 KRF81673.1 CH963913 EDW77320.2 QOIP01000008 RLU19550.1 CM002910 KMY90615.1 KMY90616.1 CH480822 EDW55515.1 EDW64517.1 KRF81672.1 CH916372 EDV99300.1 GL435766 EFN72741.1 CH902620 EDV31463.2 KPU73419.1 CH933807 EDW11507.2 PYGN01001356 PSN35459.1 CP012523 ALC39787.1 DS235093 EEB11849.1 GBGD01000954 JAC87935.1 KQ435007 KZC13581.1 APCN01005209 KQ414598 KOC69825.1 GGMR01002233 MBY14852.1 KQ435830 KOX71744.1 AXCN02000610 AEMK02000070 ABQF01040959 ABQF01040960 AL109921 KL410243 KFQ94977.1 ABLF02039202 KK646541 KFP99294.1 KK507415 KFP61848.1 KZ288348 PBC27486.1 GL452292 EFN77592.1 AKCR02000021 PKK27128.1 PCTP01000243 PIP78792.1 KK627340 KFV53840.1 MUZQ01000049 OWK60936.1 IACJ01062636 LAA46130.1 CDGI01000329 CEI96564.1 UZAE01013853 VDO11597.1 PJQM01000314 RCI05740.1 LR027517 VCU53287.1 CP011232 AKI96775.1 CP001146 NZZV01000051 MAT04280.1 LBMM01001317 KMQ96518.1 KB320813 ELW62478.1 RFIA01000243 RMF78821.1 RFIK01000126 RMF34334.1 AAOW01000023 EAR60127.1 PGTH01000016 PJF40956.1 PBFB01000029 MBJ12358.1 AP017920 BAW01065.1 CP001251 X70943 AP008226 AE017221
BABH01029075 BABH01029076 KQ460883 KPJ11239.1 JTDY01005408 KOB67042.1 AGBW02014600 OWR41357.1 KQ459586 KPI98047.1 ODYU01013276 SOQ59983.1 KQ971343 EFA03147.2 GBYB01006062 JAG75829.1 GBYB01013753 JAG83520.1 GECL01001117 JAP05007.1 GBXI01011403 JAD02889.1 GBXI01007502 JAD06790.1 GL768788 EFZ11007.1 GEDC01029607 JAS07691.1 GEDC01026550 JAS10748.1 CM000158 EDW90219.2 CH954179 EDV56668.2 AE014134 AY119180 AAF53577.3 AAM51040.1 AGB93054.1 AGB93055.1 KQ981004 KYN10393.1 CH940649 KRF81673.1 CH963913 EDW77320.2 QOIP01000008 RLU19550.1 CM002910 KMY90615.1 KMY90616.1 CH480822 EDW55515.1 EDW64517.1 KRF81672.1 CH916372 EDV99300.1 GL435766 EFN72741.1 CH902620 EDV31463.2 KPU73419.1 CH933807 EDW11507.2 PYGN01001356 PSN35459.1 CP012523 ALC39787.1 DS235093 EEB11849.1 GBGD01000954 JAC87935.1 KQ435007 KZC13581.1 APCN01005209 KQ414598 KOC69825.1 GGMR01002233 MBY14852.1 KQ435830 KOX71744.1 AXCN02000610 AEMK02000070 ABQF01040959 ABQF01040960 AL109921 KL410243 KFQ94977.1 ABLF02039202 KK646541 KFP99294.1 KK507415 KFP61848.1 KZ288348 PBC27486.1 GL452292 EFN77592.1 AKCR02000021 PKK27128.1 PCTP01000243 PIP78792.1 KK627340 KFV53840.1 MUZQ01000049 OWK60936.1 IACJ01062636 LAA46130.1 CDGI01000329 CEI96564.1 UZAE01013853 VDO11597.1 PJQM01000314 RCI05740.1 LR027517 VCU53287.1 CP011232 AKI96775.1 CP001146 NZZV01000051 MAT04280.1 LBMM01001317 KMQ96518.1 KB320813 ELW62478.1 RFIA01000243 RMF78821.1 RFIK01000126 RMF34334.1 AAOW01000023 EAR60127.1 PGTH01000016 PJF40956.1 PBFB01000029 MBJ12358.1 AP017920 BAW01065.1 CP001251 X70943 AP008226 AE017221
Proteomes
UP000005204
UP000218220
UP000053240
UP000037510
UP000007151
UP000053268
+ More
UP000007266 UP000192223 UP000002282 UP000008711 UP000000803 UP000078492 UP000008792 UP000007798 UP000279307 UP000001292 UP000001070 UP000000311 UP000007801 UP000009192 UP000245037 UP000092553 UP000009046 UP000076502 UP000075882 UP000075840 UP000075902 UP000075903 UP000001646 UP000261480 UP000261500 UP000261520 UP000233040 UP000053825 UP000053105 UP000075886 UP000008227 UP000007754 UP000005640 UP000053283 UP000007819 UP000005226 UP000005203 UP000242457 UP000245300 UP000261620 UP000079169 UP000008237 UP000076407 UP000075884 UP000261540 UP000286641 UP000053872 UP000197619 UP000038169 UP000046398 UP000278807 UP000253551 UP000279841 UP000035159 UP000001733 UP000036681 UP000036403 UP000038045 UP000011518 UP000269435 UP000002171 UP000217909 UP000007719 UP000000532 UP000000592
UP000007266 UP000192223 UP000002282 UP000008711 UP000000803 UP000078492 UP000008792 UP000007798 UP000279307 UP000001292 UP000001070 UP000000311 UP000007801 UP000009192 UP000245037 UP000092553 UP000009046 UP000076502 UP000075882 UP000075840 UP000075902 UP000075903 UP000001646 UP000261480 UP000261500 UP000261520 UP000233040 UP000053825 UP000053105 UP000075886 UP000008227 UP000007754 UP000005640 UP000053283 UP000007819 UP000005226 UP000005203 UP000242457 UP000245300 UP000261620 UP000079169 UP000008237 UP000076407 UP000075884 UP000261540 UP000286641 UP000053872 UP000197619 UP000038169 UP000046398 UP000278807 UP000253551 UP000279841 UP000035159 UP000001733 UP000036681 UP000036403 UP000038045 UP000011518 UP000269435 UP000002171 UP000217909 UP000007719 UP000000532 UP000000592
Pfam
Interpro
IPR004524
Asp-tRNA-ligase_1
+ More
IPR006195 aa-tRNA-synth_II
IPR002312 Asp/Asn-tRNA-synth_IIb
IPR004364 aa-tRNA-synt_II
IPR012340 NA-bd_OB-fold
IPR004115 GAD-like_sf
IPR004365 NA-bd_OB_tRNA
IPR035979 RBD_domain_sf
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR006535 HnRNP_R/Q_splicing_fac
IPR018149 Lys-tRNA-synth_II_C
IPR029351 GAD_dom
IPR008576 MeTrfase_NTM1
IPR029063 SAM-dependent_MTases
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR006195 aa-tRNA-synth_II
IPR002312 Asp/Asn-tRNA-synth_IIb
IPR004364 aa-tRNA-synt_II
IPR012340 NA-bd_OB-fold
IPR004115 GAD-like_sf
IPR004365 NA-bd_OB_tRNA
IPR035979 RBD_domain_sf
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR006535 HnRNP_R/Q_splicing_fac
IPR018149 Lys-tRNA-synth_II_C
IPR029351 GAD_dom
IPR008576 MeTrfase_NTM1
IPR029063 SAM-dependent_MTases
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
SUPFAM
Gene 3D
ProteinModelPortal
H9JMW1
A0A2W1BQF6
A0A2A4JII1
H9JMW0
A0A194R149
A0A0L7KUU8
+ More
A0A212EIN8 A0A194PXJ4 A0A2H1X3S0 D6WJB0 A0A0C9QZB7 A0A0C9QGP4 A0A1W4WKH8 A0A0V0GC92 A0A0A1WV06 A0A0A1X7W5 E9J836 A0A1B6C2E3 A0A1B6CB86 B4P990 B3NNL7 Q9VJH2 A0A151ITX3 A0A0Q9WAC6 B4MYY1 A0A3L8DGI4 A0A0J9R3K6 B4I559 B4LQX8 B4JQX0 E2A1R8 B3MK45 B4KJR2 A0A2P8XTY3 A0A0M4ERI2 E0VEP3 A0A069DVL5 A0A154PNX3 A0A182L2D1 A0A182IEL3 A0A182TP04 A0A182VIH7 G1KR37 A0A3B3YKL2 R4G9Q0 A0A3B3VMQ4 A0A3B4B1L7 A0A2K5R7R8 A0A0L7RG52 A0A2S2ND19 A0A0N0U498 A0A182QZW8 F1S742 A0A286ZWZ5 H1A3Z9 A0A3B3IT01 A0A091UX00 A0A2K5R7R5 J9K6D5 H2SYM8 A0A091NXS1 A0A091LQU4 A0A087ZV42 A0A2A3E8F3 S4RXF5 A0A3Q3XHH8 A0A3Q0IWP3 E2C360 A0A182WTH7 A0A182NLM4 A0A3B3RWS5 A0A3Q3XMC5 A0A3B3IS01 A0A3Q7PIA3 A0A2I0MBS3 A0A2H0D9F6 A0A093FBC9 A0A218V4N5 A0A2D4FF60 A0A0A1NXR9 A0A0R3TVN0 A0A367KUF4 A0A3P4AQ79 A0A0G2Z583 B5YDT0 A0A2E0RP97 A0A0M3I244 A0A0J7L1N1 A0A0N4ZSP0 L9KIG0 A0A3M1QHE6 A0A3M1LUE7 Q2BI63 A0A2M8PTS7 A0A2E5XYG8 A0A1J1EN87 B8E1C0 P36419 Q5SKD2 Q72KH6
A0A212EIN8 A0A194PXJ4 A0A2H1X3S0 D6WJB0 A0A0C9QZB7 A0A0C9QGP4 A0A1W4WKH8 A0A0V0GC92 A0A0A1WV06 A0A0A1X7W5 E9J836 A0A1B6C2E3 A0A1B6CB86 B4P990 B3NNL7 Q9VJH2 A0A151ITX3 A0A0Q9WAC6 B4MYY1 A0A3L8DGI4 A0A0J9R3K6 B4I559 B4LQX8 B4JQX0 E2A1R8 B3MK45 B4KJR2 A0A2P8XTY3 A0A0M4ERI2 E0VEP3 A0A069DVL5 A0A154PNX3 A0A182L2D1 A0A182IEL3 A0A182TP04 A0A182VIH7 G1KR37 A0A3B3YKL2 R4G9Q0 A0A3B3VMQ4 A0A3B4B1L7 A0A2K5R7R8 A0A0L7RG52 A0A2S2ND19 A0A0N0U498 A0A182QZW8 F1S742 A0A286ZWZ5 H1A3Z9 A0A3B3IT01 A0A091UX00 A0A2K5R7R5 J9K6D5 H2SYM8 A0A091NXS1 A0A091LQU4 A0A087ZV42 A0A2A3E8F3 S4RXF5 A0A3Q3XHH8 A0A3Q0IWP3 E2C360 A0A182WTH7 A0A182NLM4 A0A3B3RWS5 A0A3Q3XMC5 A0A3B3IS01 A0A3Q7PIA3 A0A2I0MBS3 A0A2H0D9F6 A0A093FBC9 A0A218V4N5 A0A2D4FF60 A0A0A1NXR9 A0A0R3TVN0 A0A367KUF4 A0A3P4AQ79 A0A0G2Z583 B5YDT0 A0A2E0RP97 A0A0M3I244 A0A0J7L1N1 A0A0N4ZSP0 L9KIG0 A0A3M1QHE6 A0A3M1LUE7 Q2BI63 A0A2M8PTS7 A0A2E5XYG8 A0A1J1EN87 B8E1C0 P36419 Q5SKD2 Q72KH6
PDB
4AH6
E-value=2.10723e-116,
Score=1073
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
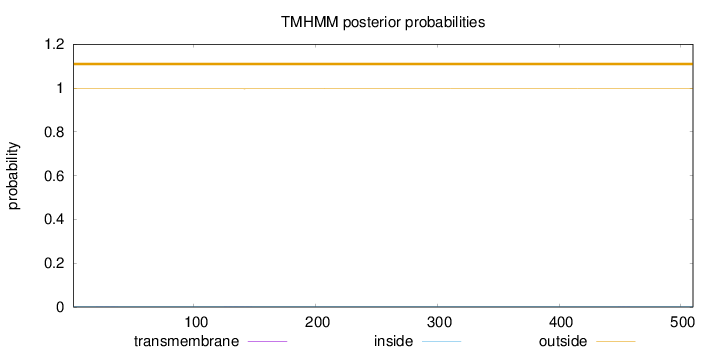
Length:
510
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0117
Exp number, first 60 AAs:
0.00374
Total prob of N-in:
0.00376
outside
1 - 510
Population Genetic Test Statistics
Pi
5.045648
Theta
3.22489
Tajima's D
-1.150948
CLR
0
CSRT
0.103694815259237
Interpretation
Uncertain
