Gene
KWMTBOMO13443
Pre Gene Modal
BGIBMGA010892
Annotation
PREDICTED:_zonadhesin-like_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.976
Sequence
CDS
ATGCCAAAGGTCACGAATGTCATCACAACACAACGCTACCGCCGCTATGTAAGAAATGATGTAGCTACTTGTCCAGAAAACGAAGAACTCTCCTGCCTTCAAAGTTTATGCAGGCCTCAGAAGTGCATCGAAGAACATGCCCCCGTGTGCCGATTAGTTGATCCGGAGATATGCGAATTTGGATGTGCCTGCAAATTAGGATATTTGAGAGACAAAAATGGAACTTGTATACCGCAAGAAAAGTGTCCAACCCAGCTTTGTTCAGTAAATGAACAGTATTTAAGTTGCATCCAAGCTGCTTGTCGGTTCGAGAAGTGTTCAGACCTGGGGGGATCCCTCAGTTGCAAGGGGGTGCCGGAAAGGGAATGCGTCGGTGGCTGCGTCTGTAAGGACAACTACTTGCGAGCTAAAAACGACACTTGTATTAAACTCAGTGATTGTGACGCTGACCTCTGCTCTGAAAATGAAATACACGTGAACTGTGTCCTAGCACAGCGTGGCCCAATGACGTGCTCAGAGAAGGACTTACTTATGCCCTACCCATTTGTGAGGCAAGAGTACTGCAAAGCCGGATGCGTGTGTAAAGAGGGATATCTGAAAGACGATAGCGGTAAATGTGTCGCCCGAGAAAACTGCCCAAATTCTGACCTCTGCTCTGAAAATGAAATACACGTGAACTGTGTCCTAGCACAGTGTGGCCCAATGACGTGCTCAGAGAAGGACTTACCTATGCCCTGCCCATTGGTGAGGCGAGAGTACTGCAAAGCCGGATGCGTGTGTAAAGAGGGATATCTGAAAGACGATAGCGGTAAATGTGTCGCCCGAGAAAACTGCCCAAATTATAGAAGAGCCGCCTAG
Protein
MPKVTNVITTQRYRRYVRNDVATCPENEELSCLQSLCRPQKCIEEHAPVCRLVDPEICEFGCACKLGYLRDKNGTCIPQEKCPTQLCSVNEQYLSCIQAACRFEKCSDLGGSLSCKGVPERECVGGCVCKDNYLRAKNDTCIKLSDCDADLCSENEIHVNCVLAQRGPMTCSEKDLLMPYPFVRQEYCKAGCVCKEGYLKDDSGKCVARENCPNSDLCSENEIHVNCVLAQCGPMTCSEKDLPMPCPLVRREYCKAGCVCKEGYLKDDSGKCVARENCPNYRRAA
Summary
Similarity
Belongs to the serpin family.
Uniprot
H9JMY9
A0A2A4IW13
A0A2A4IXH8
A0A2A4J6N9
A0A2A4J556
A0A2W1BQG2
+ More
A0A0L7L220 A0A3S2PJ20 A0A0L7L697 H9J5D8 A0A087UJV8 A0A212EGT4 A0A2H1VQQ3 A0A087TD78 A0A2A4J7B9 A0A0L7L044 A0A3G1T1Q1 A0A0V0Z7C1 A0A1E1WMT2 A0A1E1W5U8 A0A0V1EPP9 A0A2P6L3P9 A0A0V0S515 A0A2P6L3Q8 A0A194QAG0 B0XBB5 A0A194QQM3 A0A2A4J071 A0A0V1B9Y2 A0A2H1WS62 A0A3S2NJW5 A0A1J1IUR1 A0A3S2LVE8 A0A2A4JLM8 A0A0K0EAF5 A0A0V0WZ71 A0A2P6L3P8 A0A2H1VKZ4 A0A1B0GMI5 A0A3G1T1R3 A0A0V0Z7M5 G0N6T3 A0A2W1BA04 A0A0V1FGY8 A0A2P4WVB9 A0A0V1B9Z3 E3NB80 A0A0V1J3G8 A0A260ZCX5 A0A260ZCW0 A0A0V1J3E7 A0A2P4WVE3 A0A0V0Z760 A0A0V0XVX4 A0A0V1CXK4 A0A0V0Z793 A0A0V1NT84 A0A0V0VIZ3 A0A0V0U5N5 G0P062 A0A0V0U5E1 A0A2L2XYH3 A0A0V1MZF6 A0A0V0XW29 A0A0V0S5K9 A0A0V0S563 A0A0V0Z791 A0A1Y1MXL0 A0A0V1EPP1 A0A1Y1MVQ0 A0A0V1H1U7 A0A0V1EPN2 H9J5D7 A0A1I7Z0X9 A8XYE9 A0A1B0GHH0 A0A2L2XYN6 A0A1B0CE59 A0A1B0CET6 A0A194QS41 A0A194Q8M7 A0A2W1B8Z9 A0A1B0GHG8 A0A2Z6FL19 A0A2G5TUJ7 A8WLQ9 A0A2G5TTK8 A0A1J1HLL5
A0A0L7L220 A0A3S2PJ20 A0A0L7L697 H9J5D8 A0A087UJV8 A0A212EGT4 A0A2H1VQQ3 A0A087TD78 A0A2A4J7B9 A0A0L7L044 A0A3G1T1Q1 A0A0V0Z7C1 A0A1E1WMT2 A0A1E1W5U8 A0A0V1EPP9 A0A2P6L3P9 A0A0V0S515 A0A2P6L3Q8 A0A194QAG0 B0XBB5 A0A194QQM3 A0A2A4J071 A0A0V1B9Y2 A0A2H1WS62 A0A3S2NJW5 A0A1J1IUR1 A0A3S2LVE8 A0A2A4JLM8 A0A0K0EAF5 A0A0V0WZ71 A0A2P6L3P8 A0A2H1VKZ4 A0A1B0GMI5 A0A3G1T1R3 A0A0V0Z7M5 G0N6T3 A0A2W1BA04 A0A0V1FGY8 A0A2P4WVB9 A0A0V1B9Z3 E3NB80 A0A0V1J3G8 A0A260ZCX5 A0A260ZCW0 A0A0V1J3E7 A0A2P4WVE3 A0A0V0Z760 A0A0V0XVX4 A0A0V1CXK4 A0A0V0Z793 A0A0V1NT84 A0A0V0VIZ3 A0A0V0U5N5 G0P062 A0A0V0U5E1 A0A2L2XYH3 A0A0V1MZF6 A0A0V0XW29 A0A0V0S5K9 A0A0V0S563 A0A0V0Z791 A0A1Y1MXL0 A0A0V1EPP1 A0A1Y1MVQ0 A0A0V1H1U7 A0A0V1EPN2 H9J5D7 A0A1I7Z0X9 A8XYE9 A0A1B0GHH0 A0A2L2XYN6 A0A1B0CE59 A0A1B0CET6 A0A194QS41 A0A194Q8M7 A0A2W1B8Z9 A0A1B0GHG8 A0A2Z6FL19 A0A2G5TUJ7 A8WLQ9 A0A2G5TTK8 A0A1J1HLL5
Pubmed
EMBL
BABH01029075
NWSH01005913
PCG63849.1
PCG63850.1
NWSH01003013
PCG67110.1
+ More
PCG67111.1 KZ149981 PZC75815.1 JTDY01003492 KOB69475.1 RSAL01000018 RVE52843.1 JTDY01002637 KOB71038.1 BABH01039203 BABH01039204 BABH01039205 BABH01039206 BABH01039207 KK120157 KFM77647.1 AGBW02015025 OWR40695.1 ODYU01003852 SOQ43127.1 KK114672 KFM63067.1 NWSH01002542 PCG68017.1 JTDY01003897 KOB68857.1 MG770320 AXY94922.1 JYDQ01000343 KRY08317.1 GDQN01002759 JAT88295.1 GDQN01008701 JAT82353.1 JYDR01000016 KRY75718.1 MWRG01001996 PRD33208.1 JYDL01000037 KRX21785.1 PRD33209.1 KQ459249 KPJ02419.1 DS232623 EDS44206.1 KQ461181 KPJ07649.1 NWSH01004181 PCG65391.1 JYDH01000076 KRY33814.1 ODYU01010657 SOQ55910.1 RVE52846.1 CVRI01000059 CRL03442.1 RSAL01000004 RVE54612.1 NWSH01001153 PCG72343.1 JYDK01000035 KRX81073.1 PRD33210.1 ODYU01002951 SOQ41102.1 AJVK01011483 MG770321 AXY94923.1 KRY08323.1 GL379845 EGT53964.1 KZ150198 PZC72302.1 JYDT01000094 KRY85257.1 LFJK02000001 POM53504.1 KRY33812.1 DS268582 EFO91826.1 JYDS01000045 KRZ29499.1 NMWX01000177 OZF83551.1 OZF83550.1 KRZ29498.1 POM53503.1 KRY08320.1 JYDU01000118 KRX92190.1 JYDI01000077 KRY53944.1 KRY08321.1 JYDM01000104 KRZ87210.1 JYDN01000032 KRX63333.1 JYDJ01000057 KRX46475.1 GL379996 EGT41608.1 KRX46473.1 IAAA01002395 IAAA01002396 LAA01006.1 JYDO01000022 KRZ77159.1 KRX92186.1 KRX21786.1 KRX21783.1 KRY08319.1 GEZM01023997 JAV88077.1 KRY75722.1 GEZM01023999 JAV88076.1 JYDP01000166 KRZ04345.1 KRY75723.1 BABH01039220 BABH01039221 BABH01039222 BABH01039223 HE600939 CAP37666.2 AJWK01004621 AJWK01004622 IAAA01002394 LAA00998.1 AJWK01008601 AJWK01009195 AJWK01009196 KQ461175 KPJ07800.1 KQ459562 KPI99760.1 KZ150221 PZC72102.1 AJWK01004620 LC384839 BBE28982.1 PDUG01000005 PIC30636.1 HE601231 CAP21405.1 PIC30635.1 CVRI01000010 CRK88925.1
PCG67111.1 KZ149981 PZC75815.1 JTDY01003492 KOB69475.1 RSAL01000018 RVE52843.1 JTDY01002637 KOB71038.1 BABH01039203 BABH01039204 BABH01039205 BABH01039206 BABH01039207 KK120157 KFM77647.1 AGBW02015025 OWR40695.1 ODYU01003852 SOQ43127.1 KK114672 KFM63067.1 NWSH01002542 PCG68017.1 JTDY01003897 KOB68857.1 MG770320 AXY94922.1 JYDQ01000343 KRY08317.1 GDQN01002759 JAT88295.1 GDQN01008701 JAT82353.1 JYDR01000016 KRY75718.1 MWRG01001996 PRD33208.1 JYDL01000037 KRX21785.1 PRD33209.1 KQ459249 KPJ02419.1 DS232623 EDS44206.1 KQ461181 KPJ07649.1 NWSH01004181 PCG65391.1 JYDH01000076 KRY33814.1 ODYU01010657 SOQ55910.1 RVE52846.1 CVRI01000059 CRL03442.1 RSAL01000004 RVE54612.1 NWSH01001153 PCG72343.1 JYDK01000035 KRX81073.1 PRD33210.1 ODYU01002951 SOQ41102.1 AJVK01011483 MG770321 AXY94923.1 KRY08323.1 GL379845 EGT53964.1 KZ150198 PZC72302.1 JYDT01000094 KRY85257.1 LFJK02000001 POM53504.1 KRY33812.1 DS268582 EFO91826.1 JYDS01000045 KRZ29499.1 NMWX01000177 OZF83551.1 OZF83550.1 KRZ29498.1 POM53503.1 KRY08320.1 JYDU01000118 KRX92190.1 JYDI01000077 KRY53944.1 KRY08321.1 JYDM01000104 KRZ87210.1 JYDN01000032 KRX63333.1 JYDJ01000057 KRX46475.1 GL379996 EGT41608.1 KRX46473.1 IAAA01002395 IAAA01002396 LAA01006.1 JYDO01000022 KRZ77159.1 KRX92186.1 KRX21786.1 KRX21783.1 KRY08319.1 GEZM01023997 JAV88077.1 KRY75722.1 GEZM01023999 JAV88076.1 JYDP01000166 KRZ04345.1 KRY75723.1 BABH01039220 BABH01039221 BABH01039222 BABH01039223 HE600939 CAP37666.2 AJWK01004621 AJWK01004622 IAAA01002394 LAA00998.1 AJWK01008601 AJWK01009195 AJWK01009196 KQ461175 KPJ07800.1 KQ459562 KPI99760.1 KZ150221 PZC72102.1 AJWK01004620 LC384839 BBE28982.1 PDUG01000005 PIC30636.1 HE601231 CAP21405.1 PIC30635.1 CVRI01000010 CRK88925.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000283053
UP000054359
UP000007151
+ More
UP000054783 UP000054632 UP000054630 UP000053268 UP000002320 UP000053240 UP000054776 UP000183832 UP000035681 UP000054673 UP000092462 UP000008068 UP000054995 UP000237256 UP000008281 UP000054805 UP000216624 UP000054815 UP000054653 UP000054924 UP000054681 UP000055048 UP000054843 UP000055024 UP000095287 UP000008549 UP000092461 UP000230233
UP000054783 UP000054632 UP000054630 UP000053268 UP000002320 UP000053240 UP000054776 UP000183832 UP000035681 UP000054673 UP000092462 UP000008068 UP000054995 UP000237256 UP000008281 UP000054805 UP000216624 UP000054815 UP000054653 UP000054924 UP000054681 UP000055048 UP000054843 UP000055024 UP000095287 UP000008549 UP000092461 UP000230233
Pfam
Interpro
IPR036084
Ser_inhib-like_sf
+ More
IPR002919 TIL_dom
IPR004094 Antistasin-like
IPR017896 4Fe4S_Fe-S-bd
IPR042185 Serpin_sf_2
IPR023796 Serpin_dom
IPR042178 Serpin_sf_1
IPR036186 Serpin_sf
IPR000742 EGF-like_dom
IPR000215 Serpin_fam
IPR023795 Serpin_CS
IPR011236 Ser/Thr_PPase_5
IPR013235 PPP_dom
IPR016092 FeS_cluster_insertion
IPR029052 Metallo-depent_PP-like
IPR041753 PP5_C
IPR035903 HesB-like_dom_sf
IPR019734 TPR_repeat
IPR011990 TPR-like_helical_dom_sf
IPR004855 TFIIA_asu/bsu
IPR004843 Calcineurin-like_PHP_ApaH
IPR013026 TPR-contain_dom
IPR006186 Ser/Thr-sp_prot-phosphatase
IPR000361 FeS_biogenesis
IPR006212 Furin_repeat
IPR003645 Fol_N
IPR002919 TIL_dom
IPR004094 Antistasin-like
IPR017896 4Fe4S_Fe-S-bd
IPR042185 Serpin_sf_2
IPR023796 Serpin_dom
IPR042178 Serpin_sf_1
IPR036186 Serpin_sf
IPR000742 EGF-like_dom
IPR000215 Serpin_fam
IPR023795 Serpin_CS
IPR011236 Ser/Thr_PPase_5
IPR013235 PPP_dom
IPR016092 FeS_cluster_insertion
IPR029052 Metallo-depent_PP-like
IPR041753 PP5_C
IPR035903 HesB-like_dom_sf
IPR019734 TPR_repeat
IPR011990 TPR-like_helical_dom_sf
IPR004855 TFIIA_asu/bsu
IPR004843 Calcineurin-like_PHP_ApaH
IPR013026 TPR-contain_dom
IPR006186 Ser/Thr-sp_prot-phosphatase
IPR000361 FeS_biogenesis
IPR006212 Furin_repeat
IPR003645 Fol_N
ProteinModelPortal
H9JMY9
A0A2A4IW13
A0A2A4IXH8
A0A2A4J6N9
A0A2A4J556
A0A2W1BQG2
+ More
A0A0L7L220 A0A3S2PJ20 A0A0L7L697 H9J5D8 A0A087UJV8 A0A212EGT4 A0A2H1VQQ3 A0A087TD78 A0A2A4J7B9 A0A0L7L044 A0A3G1T1Q1 A0A0V0Z7C1 A0A1E1WMT2 A0A1E1W5U8 A0A0V1EPP9 A0A2P6L3P9 A0A0V0S515 A0A2P6L3Q8 A0A194QAG0 B0XBB5 A0A194QQM3 A0A2A4J071 A0A0V1B9Y2 A0A2H1WS62 A0A3S2NJW5 A0A1J1IUR1 A0A3S2LVE8 A0A2A4JLM8 A0A0K0EAF5 A0A0V0WZ71 A0A2P6L3P8 A0A2H1VKZ4 A0A1B0GMI5 A0A3G1T1R3 A0A0V0Z7M5 G0N6T3 A0A2W1BA04 A0A0V1FGY8 A0A2P4WVB9 A0A0V1B9Z3 E3NB80 A0A0V1J3G8 A0A260ZCX5 A0A260ZCW0 A0A0V1J3E7 A0A2P4WVE3 A0A0V0Z760 A0A0V0XVX4 A0A0V1CXK4 A0A0V0Z793 A0A0V1NT84 A0A0V0VIZ3 A0A0V0U5N5 G0P062 A0A0V0U5E1 A0A2L2XYH3 A0A0V1MZF6 A0A0V0XW29 A0A0V0S5K9 A0A0V0S563 A0A0V0Z791 A0A1Y1MXL0 A0A0V1EPP1 A0A1Y1MVQ0 A0A0V1H1U7 A0A0V1EPN2 H9J5D7 A0A1I7Z0X9 A8XYE9 A0A1B0GHH0 A0A2L2XYN6 A0A1B0CE59 A0A1B0CET6 A0A194QS41 A0A194Q8M7 A0A2W1B8Z9 A0A1B0GHG8 A0A2Z6FL19 A0A2G5TUJ7 A8WLQ9 A0A2G5TTK8 A0A1J1HLL5
A0A0L7L220 A0A3S2PJ20 A0A0L7L697 H9J5D8 A0A087UJV8 A0A212EGT4 A0A2H1VQQ3 A0A087TD78 A0A2A4J7B9 A0A0L7L044 A0A3G1T1Q1 A0A0V0Z7C1 A0A1E1WMT2 A0A1E1W5U8 A0A0V1EPP9 A0A2P6L3P9 A0A0V0S515 A0A2P6L3Q8 A0A194QAG0 B0XBB5 A0A194QQM3 A0A2A4J071 A0A0V1B9Y2 A0A2H1WS62 A0A3S2NJW5 A0A1J1IUR1 A0A3S2LVE8 A0A2A4JLM8 A0A0K0EAF5 A0A0V0WZ71 A0A2P6L3P8 A0A2H1VKZ4 A0A1B0GMI5 A0A3G1T1R3 A0A0V0Z7M5 G0N6T3 A0A2W1BA04 A0A0V1FGY8 A0A2P4WVB9 A0A0V1B9Z3 E3NB80 A0A0V1J3G8 A0A260ZCX5 A0A260ZCW0 A0A0V1J3E7 A0A2P4WVE3 A0A0V0Z760 A0A0V0XVX4 A0A0V1CXK4 A0A0V0Z793 A0A0V1NT84 A0A0V0VIZ3 A0A0V0U5N5 G0P062 A0A0V0U5E1 A0A2L2XYH3 A0A0V1MZF6 A0A0V0XW29 A0A0V0S5K9 A0A0V0S563 A0A0V0Z791 A0A1Y1MXL0 A0A0V1EPP1 A0A1Y1MVQ0 A0A0V1H1U7 A0A0V1EPN2 H9J5D7 A0A1I7Z0X9 A8XYE9 A0A1B0GHH0 A0A2L2XYN6 A0A1B0CE59 A0A1B0CET6 A0A194QS41 A0A194Q8M7 A0A2W1B8Z9 A0A1B0GHG8 A0A2Z6FL19 A0A2G5TUJ7 A8WLQ9 A0A2G5TTK8 A0A1J1HLL5
Ontologies
GO
PANTHER
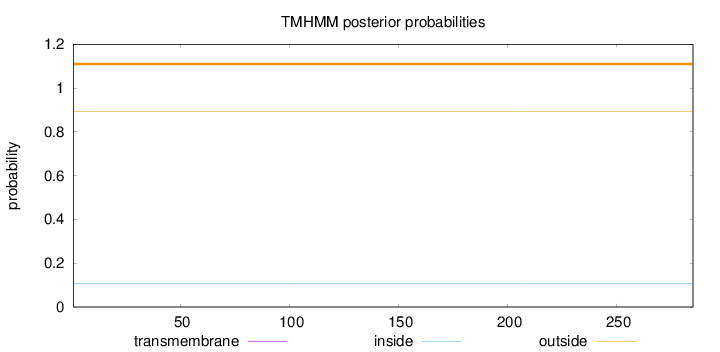
Topology
Length:
285
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10869
outside
1 - 285
Population Genetic Test Statistics
Pi
17.741687
Theta
13.767309
Tajima's D
-0.51462
CLR
2.346833
CSRT
0.24228788560572
Interpretation
Uncertain
