Gene
KWMTBOMO13440
Pre Gene Modal
BGIBMGA010861
Annotation
PREDICTED:_sialin_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.932
Sequence
CDS
ATGGACTGCATTACTTACATTGTATCAGAACAAACAAAAGACCATGAGATGACGTGGCGTTTCTGGCGACGGCGGCGTTTAGTGGTAGCGCTGTTGGCATTCTTCGGGTTCTTCAACGTTTACGCTCTTCGCGTGAACCTCTCAGTGGCCGTCGTGGCCATGACGGAACCGGTGGAAACGAAACTGGAAAACGGTACCATTGCTATGATTCCAGAATTCGACTGGAGCACGCAAACGAAAGGCCTCGTCCTAAGTTCGTTTTTCTACGGATACCTAGTCACTCAGCTCCCTGGCGGCTGGCTCGCTGCTAAAATAGGAGGTAACAGGGTGTTCGCTATCGGCATCGGGACCACGTCGCTGCTAACCCTGTTCACGCCTCCGCTCGCCCACACCAGCACGGCGCTACTCATCGCCGTCAGAGTTATCGAAGGATTCTTCGAGGGCGTGACGTACCCGTGCATCCACGCGGTGTGGGCGCGCTGGGCGCCGGGCGAGGAGCGCGCGCGGCTGGCCACGCTCGCCTTCAGCGGCAGCTACGCCGGCACCGTGGTGGCCATGCCGCTGTGCTCGCTGCTGGCGCACTACACGGGCTGGCCCGGCATCTTCTACGTCTTCGGTATACTGGGATTGATTTGGACGACAGTGTGGTGGATAGTTGTGAAGGAGTCCCCGGAGAAGGACCCAAGGATTAGCGCCGCAGAACTCAAGTACATACAGGAGACTCGAGGCACGACGCTGACTGATGGAGCGAAGCACCCCTGGAAGGCGATGCTGACGTCGGGTCCGGTGTGGGCCATTGTGGTGGCGCACTTCAGCGAGAACTGGGGCTTCTATACTTTACTCACGTTCTTGCCTACGTTCATGAAAGATGTGTACAAGTTCGAGACGTCGGCCACGGGCTGGCTGTCGGCCATCCCGTACCTGGCGATGGCGGCCACGCTGCAGGGCGCCGGACACCTGGCCGACTGCCTGCTGCGCCGCGGCCTGCTGTCCCGGACCAACATACGGAAGCTGTTCAACTGCGGAGCCTTCCTGTCCCAGACCGTGTTCATGATAGCGGCGGCCTACTCCACGTCGGTGGTGGGCTGCGTCGTGTGCCTGACTATAGCCGTCGGCCTGGGGGGCTTCGCGTGGTCCGGCTTCAGCGTGAACCACCTGGACATCGCGCCGCCCCACGCCAGCGTGCTGATGGGCGTCTCCAACACGTTCGCCACGCTGCCCGGGATCGTGAGCCCGCCCCTCGCCGGGAGCATCGTGCTCGACAAGACGGCGGAGCAATGGCGCGTCGTGTTCTTCATATCGAGCGCGATCTATCTGTTCGGCGCCGTCGTGTACTACATATTCTGCTCGGCCGAGCTGCAGCCCTGGGTGCTGCACGACCAGTCCGACGCCAGCTTCGACACGGACGGCGCCTCCGTCACCACGGCCGGCGGATACGACAACAGGGCCACGGACCACAACTCGGAAATGTAA
Protein
MDCITYIVSEQTKDHEMTWRFWRRRRLVVALLAFFGFFNVYALRVNLSVAVVAMTEPVETKLENGTIAMIPEFDWSTQTKGLVLSSFFYGYLVTQLPGGWLAAKIGGNRVFAIGIGTTSLLTLFTPPLAHTSTALLIAVRVIEGFFEGVTYPCIHAVWARWAPGEERARLATLAFSGSYAGTVVAMPLCSLLAHYTGWPGIFYVFGILGLIWTTVWWIVVKESPEKDPRISAAELKYIQETRGTTLTDGAKHPWKAMLTSGPVWAIVVAHFSENWGFYTLLTFLPTFMKDVYKFETSATGWLSAIPYLAMAATLQGAGHLADCLLRRGLLSRTNIRKLFNCGAFLSQTVFMIAAAYSTSVVGCVVCLTIAVGLGGFAWSGFSVNHLDIAPPHASVLMGVSNTFATLPGIVSPPLAGSIVLDKTAEQWRVVFFISSAIYLFGAVVYYIFCSAELQPWVLHDQSDASFDTDGASVTTAGGYDNRATDHNSEM
Summary
Uniprot
H9JMV8
A0A2H1V7V5
A0A2W1BL51
A0A194Q3Q7
A0A212EIP7
A0A2J7QA10
+ More
D6WN85 A0A1Y1KE92 A0A0T6B3N9 A0A023F873 A0A224XPJ3 T1IC35 R4G7U5 A0A310SHD6 A0A088AQK9 A0A1B6KF51 A0A2A3EH07 A0A067R451 A0A1Y1L7V4 A0A2J7QA17 A0A1L8DEF1 A0A1L8DEA4 A0A1S4N2F7 A0A1W4XUW2 A0A1W4XVT2 A0A1W4XVW4 E0W365 A0A232FI30 A0A0L0BTU3 A0A154NX58 A0A1I8Q9J5 V5GQF3 A0A023ET50 A0A1Y1MZH0 A0A182GVH0 A0A182GDT7 Q17DB4 A0A023EU55 A0A1A9W4V1 B4G3D7 A0A1A9XSF6 A0A1B0BRR3 A0A1L8ECX5 A0A1A9Z960 Q298B7 A0A1W4XKR8 A0A1A9VHC3 A0A1L8DE89 A0A067RAW1 A0A1B0FB71 A0A1L8DES6 B3MSN7 A0A1I8MHE4 B0WK19 A0A0A1X9S1 A0A3B0K3R5 A0A1Q3F7G4 A0A1Q3F7L6 A0A2M4BLT2 A0A034VK59 B4QS29 A0A182X4H4 W8CBH9 B3NYY2 A0A1Y1LXB3 A0A2M4APS9 W5J4E4 A0A182PMT2 A0A182UMK9 A0A182LKF0 Q7Q580 A0A182I4H1 A0A1L8DE27 A0A1L8DEA2 B4NFX5 A0A1W4UXR0 A0A1L8DEK7 A0A2M4AQT8 A0A1L8DEB5 A0A182QGD0 A0A182FEW8 A0A2M3Z947 A0A182K0C0 A0A084WGH0 Q7K2N3 A0A182NIB1 E0VT50 B4PP96 B4K7S0 A0A0M4EGJ3 A0A1I8Q9P3 A0A182R943 A0A182WBS0 A0A1Y1JTQ6 A0A2R7W2R0 A0A182Y2L2 K7J9C8 B4M1A5 A0A1J1J6W1
D6WN85 A0A1Y1KE92 A0A0T6B3N9 A0A023F873 A0A224XPJ3 T1IC35 R4G7U5 A0A310SHD6 A0A088AQK9 A0A1B6KF51 A0A2A3EH07 A0A067R451 A0A1Y1L7V4 A0A2J7QA17 A0A1L8DEF1 A0A1L8DEA4 A0A1S4N2F7 A0A1W4XUW2 A0A1W4XVT2 A0A1W4XVW4 E0W365 A0A232FI30 A0A0L0BTU3 A0A154NX58 A0A1I8Q9J5 V5GQF3 A0A023ET50 A0A1Y1MZH0 A0A182GVH0 A0A182GDT7 Q17DB4 A0A023EU55 A0A1A9W4V1 B4G3D7 A0A1A9XSF6 A0A1B0BRR3 A0A1L8ECX5 A0A1A9Z960 Q298B7 A0A1W4XKR8 A0A1A9VHC3 A0A1L8DE89 A0A067RAW1 A0A1B0FB71 A0A1L8DES6 B3MSN7 A0A1I8MHE4 B0WK19 A0A0A1X9S1 A0A3B0K3R5 A0A1Q3F7G4 A0A1Q3F7L6 A0A2M4BLT2 A0A034VK59 B4QS29 A0A182X4H4 W8CBH9 B3NYY2 A0A1Y1LXB3 A0A2M4APS9 W5J4E4 A0A182PMT2 A0A182UMK9 A0A182LKF0 Q7Q580 A0A182I4H1 A0A1L8DE27 A0A1L8DEA2 B4NFX5 A0A1W4UXR0 A0A1L8DEK7 A0A2M4AQT8 A0A1L8DEB5 A0A182QGD0 A0A182FEW8 A0A2M3Z947 A0A182K0C0 A0A084WGH0 Q7K2N3 A0A182NIB1 E0VT50 B4PP96 B4K7S0 A0A0M4EGJ3 A0A1I8Q9P3 A0A182R943 A0A182WBS0 A0A1Y1JTQ6 A0A2R7W2R0 A0A182Y2L2 K7J9C8 B4M1A5 A0A1J1J6W1
Pubmed
19121390
28756777
26354079
22118469
18362917
19820115
+ More
28004739 25474469 24845553 20566863 28648823 26108605 24945155 26483478 17510324 17994087 15632085 25315136 25830018 25348373 24495485 20920257 23761445 20966253 12364791 14747013 17210077 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25244985 20075255
28004739 25474469 24845553 20566863 28648823 26108605 24945155 26483478 17510324 17994087 15632085 25315136 25830018 25348373 24495485 20920257 23761445 20966253 12364791 14747013 17210077 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25244985 20075255
EMBL
BABH01029077
BABH01029078
BABH01029079
ODYU01001137
SOQ36928.1
KZ149981
+ More
PZC75802.1 KQ459586 KPI98045.1 AGBW02014600 OWR41359.1 NEVH01016339 PNF25428.1 KQ971342 EFA03048.1 GEZM01087709 JAV58520.1 LJIG01016002 KRT81910.1 GBBI01001267 JAC17445.1 GFTR01006485 JAW09941.1 ACPB03017885 GAHY01002112 JAA75398.1 KQ769580 OAD52852.1 GEBQ01029900 JAT10077.1 KZ288253 PBC31017.1 KK852994 KDR12727.1 GEZM01063985 JAV68878.1 PNF25426.1 GFDF01009319 JAV04765.1 GFDF01009302 JAV04782.1 AAZO01007334 DS235882 EEB20071.1 NNAY01000193 OXU30108.1 JRES01001347 KNC23500.1 KQ434777 KZC04249.1 GALX01002092 JAB66374.1 GAPW01001140 JAC12458.1 GEZM01016925 GEZM01016924 JAV91044.1 JXUM01018920 JXUM01018921 KQ560526 KXJ81973.1 JXUM01016752 JXUM01016753 JXUM01016754 JXUM01016755 JXUM01016756 CH477297 EAT44376.1 GAPW01001141 JAC12457.1 CH479179 EDW24319.1 JXJN01019252 JXJN01019253 GFDG01002376 JAV16423.1 CM000070 EAL28038.1 GFDF01009318 JAV04766.1 KK852584 KDR20852.1 CCAG010022183 GFDF01009234 JAV04850.1 CH902623 EDV30277.1 DS231966 EDS29563.1 GBXI01012456 GBXI01010704 GBXI01006606 GBXI01001095 JAD01836.1 JAD03588.1 JAD07686.1 JAD13197.1 OUUW01000007 SPP82590.1 GFDL01011534 JAV23511.1 GFDL01011481 JAV23564.1 GGFJ01004899 MBW54040.1 GAKP01016078 GAKP01016077 JAC42874.1 CM000364 EDX12225.1 GAMC01004776 JAC01780.1 CH954181 EDV48385.1 GEZM01044474 JAV78192.1 GGFK01009474 MBW42795.1 ADMH02002146 ETN58283.1 AAAB01008960 EAA11677.3 APCN01002312 GFDF01009378 JAV04706.1 GFDF01009377 JAV04707.1 CH964251 EDW83192.1 GFDF01009269 JAV04815.1 GGFK01009818 MBW43139.1 GFDF01009306 JAV04778.1 AXCN02000879 GGFM01004197 MBW24948.1 ATLV01023569 KE525344 KFB49314.1 AE014297 AY060776 AAF55770.1 AAF55771.1 AAL28324.1 DS235759 EEB16556.1 CM000160 EDW96135.2 CH933806 EDW16441.1 CP012526 ALC47504.1 GEZM01101296 JAV52699.1 KK854272 PTY14044.1 CH940650 EDW67516.1 CVRI01000067 CRL06625.1
PZC75802.1 KQ459586 KPI98045.1 AGBW02014600 OWR41359.1 NEVH01016339 PNF25428.1 KQ971342 EFA03048.1 GEZM01087709 JAV58520.1 LJIG01016002 KRT81910.1 GBBI01001267 JAC17445.1 GFTR01006485 JAW09941.1 ACPB03017885 GAHY01002112 JAA75398.1 KQ769580 OAD52852.1 GEBQ01029900 JAT10077.1 KZ288253 PBC31017.1 KK852994 KDR12727.1 GEZM01063985 JAV68878.1 PNF25426.1 GFDF01009319 JAV04765.1 GFDF01009302 JAV04782.1 AAZO01007334 DS235882 EEB20071.1 NNAY01000193 OXU30108.1 JRES01001347 KNC23500.1 KQ434777 KZC04249.1 GALX01002092 JAB66374.1 GAPW01001140 JAC12458.1 GEZM01016925 GEZM01016924 JAV91044.1 JXUM01018920 JXUM01018921 KQ560526 KXJ81973.1 JXUM01016752 JXUM01016753 JXUM01016754 JXUM01016755 JXUM01016756 CH477297 EAT44376.1 GAPW01001141 JAC12457.1 CH479179 EDW24319.1 JXJN01019252 JXJN01019253 GFDG01002376 JAV16423.1 CM000070 EAL28038.1 GFDF01009318 JAV04766.1 KK852584 KDR20852.1 CCAG010022183 GFDF01009234 JAV04850.1 CH902623 EDV30277.1 DS231966 EDS29563.1 GBXI01012456 GBXI01010704 GBXI01006606 GBXI01001095 JAD01836.1 JAD03588.1 JAD07686.1 JAD13197.1 OUUW01000007 SPP82590.1 GFDL01011534 JAV23511.1 GFDL01011481 JAV23564.1 GGFJ01004899 MBW54040.1 GAKP01016078 GAKP01016077 JAC42874.1 CM000364 EDX12225.1 GAMC01004776 JAC01780.1 CH954181 EDV48385.1 GEZM01044474 JAV78192.1 GGFK01009474 MBW42795.1 ADMH02002146 ETN58283.1 AAAB01008960 EAA11677.3 APCN01002312 GFDF01009378 JAV04706.1 GFDF01009377 JAV04707.1 CH964251 EDW83192.1 GFDF01009269 JAV04815.1 GGFK01009818 MBW43139.1 GFDF01009306 JAV04778.1 AXCN02000879 GGFM01004197 MBW24948.1 ATLV01023569 KE525344 KFB49314.1 AE014297 AY060776 AAF55770.1 AAF55771.1 AAL28324.1 DS235759 EEB16556.1 CM000160 EDW96135.2 CH933806 EDW16441.1 CP012526 ALC47504.1 GEZM01101296 JAV52699.1 KK854272 PTY14044.1 CH940650 EDW67516.1 CVRI01000067 CRL06625.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000235965
UP000007266
UP000015103
+ More
UP000005203 UP000242457 UP000027135 UP000192223 UP000009046 UP000215335 UP000037069 UP000076502 UP000095300 UP000069940 UP000249989 UP000008820 UP000091820 UP000008744 UP000092443 UP000092460 UP000092445 UP000001819 UP000078200 UP000092444 UP000007801 UP000095301 UP000002320 UP000268350 UP000000304 UP000076407 UP000008711 UP000000673 UP000075885 UP000075903 UP000075882 UP000007062 UP000075840 UP000007798 UP000192221 UP000075886 UP000069272 UP000075881 UP000030765 UP000000803 UP000075884 UP000002282 UP000009192 UP000092553 UP000075900 UP000075920 UP000076408 UP000002358 UP000008792 UP000183832
UP000005203 UP000242457 UP000027135 UP000192223 UP000009046 UP000215335 UP000037069 UP000076502 UP000095300 UP000069940 UP000249989 UP000008820 UP000091820 UP000008744 UP000092443 UP000092460 UP000092445 UP000001819 UP000078200 UP000092444 UP000007801 UP000095301 UP000002320 UP000268350 UP000000304 UP000076407 UP000008711 UP000000673 UP000075885 UP000075903 UP000075882 UP000007062 UP000075840 UP000007798 UP000192221 UP000075886 UP000069272 UP000075881 UP000030765 UP000000803 UP000075884 UP000002282 UP000009192 UP000092553 UP000075900 UP000075920 UP000076408 UP000002358 UP000008792 UP000183832
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JMV8
A0A2H1V7V5
A0A2W1BL51
A0A194Q3Q7
A0A212EIP7
A0A2J7QA10
+ More
D6WN85 A0A1Y1KE92 A0A0T6B3N9 A0A023F873 A0A224XPJ3 T1IC35 R4G7U5 A0A310SHD6 A0A088AQK9 A0A1B6KF51 A0A2A3EH07 A0A067R451 A0A1Y1L7V4 A0A2J7QA17 A0A1L8DEF1 A0A1L8DEA4 A0A1S4N2F7 A0A1W4XUW2 A0A1W4XVT2 A0A1W4XVW4 E0W365 A0A232FI30 A0A0L0BTU3 A0A154NX58 A0A1I8Q9J5 V5GQF3 A0A023ET50 A0A1Y1MZH0 A0A182GVH0 A0A182GDT7 Q17DB4 A0A023EU55 A0A1A9W4V1 B4G3D7 A0A1A9XSF6 A0A1B0BRR3 A0A1L8ECX5 A0A1A9Z960 Q298B7 A0A1W4XKR8 A0A1A9VHC3 A0A1L8DE89 A0A067RAW1 A0A1B0FB71 A0A1L8DES6 B3MSN7 A0A1I8MHE4 B0WK19 A0A0A1X9S1 A0A3B0K3R5 A0A1Q3F7G4 A0A1Q3F7L6 A0A2M4BLT2 A0A034VK59 B4QS29 A0A182X4H4 W8CBH9 B3NYY2 A0A1Y1LXB3 A0A2M4APS9 W5J4E4 A0A182PMT2 A0A182UMK9 A0A182LKF0 Q7Q580 A0A182I4H1 A0A1L8DE27 A0A1L8DEA2 B4NFX5 A0A1W4UXR0 A0A1L8DEK7 A0A2M4AQT8 A0A1L8DEB5 A0A182QGD0 A0A182FEW8 A0A2M3Z947 A0A182K0C0 A0A084WGH0 Q7K2N3 A0A182NIB1 E0VT50 B4PP96 B4K7S0 A0A0M4EGJ3 A0A1I8Q9P3 A0A182R943 A0A182WBS0 A0A1Y1JTQ6 A0A2R7W2R0 A0A182Y2L2 K7J9C8 B4M1A5 A0A1J1J6W1
D6WN85 A0A1Y1KE92 A0A0T6B3N9 A0A023F873 A0A224XPJ3 T1IC35 R4G7U5 A0A310SHD6 A0A088AQK9 A0A1B6KF51 A0A2A3EH07 A0A067R451 A0A1Y1L7V4 A0A2J7QA17 A0A1L8DEF1 A0A1L8DEA4 A0A1S4N2F7 A0A1W4XUW2 A0A1W4XVT2 A0A1W4XVW4 E0W365 A0A232FI30 A0A0L0BTU3 A0A154NX58 A0A1I8Q9J5 V5GQF3 A0A023ET50 A0A1Y1MZH0 A0A182GVH0 A0A182GDT7 Q17DB4 A0A023EU55 A0A1A9W4V1 B4G3D7 A0A1A9XSF6 A0A1B0BRR3 A0A1L8ECX5 A0A1A9Z960 Q298B7 A0A1W4XKR8 A0A1A9VHC3 A0A1L8DE89 A0A067RAW1 A0A1B0FB71 A0A1L8DES6 B3MSN7 A0A1I8MHE4 B0WK19 A0A0A1X9S1 A0A3B0K3R5 A0A1Q3F7G4 A0A1Q3F7L6 A0A2M4BLT2 A0A034VK59 B4QS29 A0A182X4H4 W8CBH9 B3NYY2 A0A1Y1LXB3 A0A2M4APS9 W5J4E4 A0A182PMT2 A0A182UMK9 A0A182LKF0 Q7Q580 A0A182I4H1 A0A1L8DE27 A0A1L8DEA2 B4NFX5 A0A1W4UXR0 A0A1L8DEK7 A0A2M4AQT8 A0A1L8DEB5 A0A182QGD0 A0A182FEW8 A0A2M3Z947 A0A182K0C0 A0A084WGH0 Q7K2N3 A0A182NIB1 E0VT50 B4PP96 B4K7S0 A0A0M4EGJ3 A0A1I8Q9P3 A0A182R943 A0A182WBS0 A0A1Y1JTQ6 A0A2R7W2R0 A0A182Y2L2 K7J9C8 B4M1A5 A0A1J1J6W1
PDB
6E9N
E-value=5.2433e-22,
Score=259
Ontologies
GO
Topology
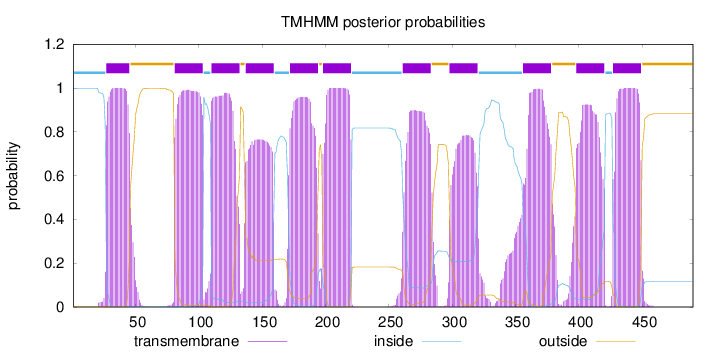
Length:
490
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
231.04859
Exp number, first 60 AAs:
20.38098
Total prob of N-in:
0.99856
POSSIBLE N-term signal
sequence
inside
1 - 26
TMhelix
27 - 45
outside
46 - 80
TMhelix
81 - 103
inside
104 - 109
TMhelix
110 - 132
outside
133 - 136
TMhelix
137 - 159
inside
160 - 171
TMhelix
172 - 194
outside
195 - 197
TMhelix
198 - 220
inside
221 - 260
TMhelix
261 - 283
outside
284 - 297
TMhelix
298 - 320
inside
321 - 355
TMhelix
356 - 378
outside
379 - 397
TMhelix
398 - 420
inside
421 - 426
TMhelix
427 - 449
outside
450 - 490
Population Genetic Test Statistics
Pi
22.175204
Theta
23.103422
Tajima's D
-1.085036
CLR
10.709689
CSRT
0.125443727813609
Interpretation
Uncertain
