Gene
KWMTBOMO13437
Pre Gene Modal
BGIBMGA014309
Annotation
neuropeptide_receptor_A26_[Bombyx_mori]
Full name
Cardioacceleratory peptide receptor
Alternative Name
Crustacean cardioactive peptide receptor
Location in the cell
PlasmaMembrane Reliability : 4.985
Sequence
CDS
ATGCCAAAATATGATTTCCTGATATTGTTACCATACCAAATACCAGATTTAGATTTATTCGTGGGTTTGACCTACGTGTTCCCTGATATATTACAGAAGATCATCATTGCTTGGTACGCGGGCGAGTTTATGTGCAAAACCGTCAAATTCCTGCAGGCCGTTGTGATGTACGCGTCGACGTACGTGCTGGTCGCGTTGAGTATCGACCGTTGCGACGCCATCACAAACCCCATGAACTTCTCCGGGAGCTGGAATCGAGCGCGGGTCCTCGTCGTGTCCGCCTGGCTAATCAGCGTAATATTCTCCATTCCGCTTTTCATCTTATACGAAGTCAAAGAAGTTCAAGGTGAACTCCAATGCTGGATCGATTTGGGAAACCCAAAGCGATGGCGCATCTGGGTGACTCTGGTGTCCATGATGATATTCATCCTTCCTGCCTTGACCATAGCTGCATGTTACGCGGTCATCGTGCTCACGATTTGGACCAAAAGCAAGGCTGTAGTGATGAGCCCTCCGATTTCTTCCAGGAGGACCAAGACTATGAGGAATGGTCAGATAGAGAGCGACCCGGATTCGAGGAGGGCCAGCTCCCGCGGACTGATACCAAGAGCCAAGATCAAGAGCGTCAAAATGACATTCGTCATTGTTTTCGTGTTCGTATTGTGCTGGTCTCCGTATATAGTGTTCGATCTGCTCCAAGTGTACGGTCACATTCCGTCGACGCAGCACTACTCCGCCATCGCCACCCTCATCCAGAGCCTCGCGCCCCTCAACTCGGCCGCCAATCCTCTTATTTGCTGCATGTTCTCTCCATACATTTACACTAGTTTGCGACGCGTGCCCCCCTACAAGTGGATATGGTGGTTGGGTCGCCATAAGCGCGCGGGCCGCGGCACACTCCGGTCCCGCTCAGACTCCACAGCCCACTCCGACCTGCTAAGCTCCACGCACGCACGACGCTCGCACTCTGTAGCAACGTTCTCATTTGTGGCACTGTCGTTCCAATGTGATGAGTGA
Protein
MPKYDFLILLPYQIPDLDLFVGLTYVFPDILQKIIIAWYAGEFMCKTVKFLQAVVMYASTYVLVALSIDRCDAITNPMNFSGSWNRARVLVVSAWLISVIFSIPLFILYEVKEVQGELQCWIDLGNPKRWRIWVTLVSMMIFILPALTIAACYAVIVLTIWTKSKAVVMSPPISSRRTKTMRNGQIESDPDSRRASSRGLIPRAKIKSVKMTFVIVFVFVLCWSPYIVFDLLQVYGHIPSTQHYSAIATLIQSLAPLNSAANPLICCMFSPYIYTSLRRVPPYKWIWWLGRHKRAGRGTLRSRSDSTAHSDLLSSTHARRSHSVATFSFVALSFQCDE
Summary
Description
Binds to the cardioactive peptide (CCAP), which is a neuropeptide.
Similarity
Belongs to the G-protein coupled receptor 1 family. Vasopressin/oxytocin receptor subfamily.
Keywords
Cell membrane
Complete proteome
G-protein coupled receptor
Glycoprotein
Membrane
Receptor
Reference proteome
Transducer
Transmembrane
Transmembrane helix
Feature
chain Cardioacceleratory peptide receptor
Uniprot
B3XXN9
A0A2H1VI11
A0A2W1BNF2
A0A194PYE0
A0A0S1YDW4
A0A212F010
+ More
A0A0N1ID10 A0A194PXI9 A0A0N1PHB6 A0A2A4JVC9 A0A2W1BIG4 B3XXP3 A0A3S2NWT7 D6WNN7 A0A0S1YDI1 A0A023F8Q5 A0A1Y1LM32 A3RE79 A0A139WI74 T1HB42 A0A2J7QPH5 A3RE80 U3UA05 K9JB71 E2AMN8 S5XYH3 A0A1B6MMH0 A0A3L8DHW8 E2BEV4 A0A1I8NBB0 N1NSV9 A0A026VWL4 A0A182XBF7 A0A0P5DAL8 A0A182F845 A0A0K8WE96 A0A0A1XTA7 A0A0P5XV02 A0A182VYW8 A0A212EZY2 A0A0P5DKE7 A0A182V4S4 A0A0P5UCK7 A0A182I098 A0A0P5D3F6 A0A0P5VKY2 A0A0N8EQ24 B0WG36 A0A0P6BPA6 A0A034WA99 A0A182UA01 Q7PYI9 A0A182RPX1 A0A182Y5B5 A0A1I8P294 A0A1A9UN78 U3U4F2 A0A182PVL2 A0A067RFL8 A0A0P5S7T5 A0A1B0G6W9 Q1RKW4 A0A0R1E5S3 A0A0P5S165 A0A0P5D985 A0A182F844 A0A1B0BRH7 A0A1A9YK95 A0A182LU62 A0A182NS99 A0A0P5IEN0 B3LYJ1 A0A3B0K5U2 A0A182PZC5 A0A1B0AAA9 B4QVZ9 A0A182MEC2 A0A1A9W8A8 A0A182I097 B4K6B4 X1WN36 B5DX60 A0A182U3G5 A0A182XBF6 B4GEI1 B4NJA1 A0A2A3EK67 H5V8A1 A0A182KLW8 A0A182PVL3 F4WNW6 Q868T3 A0A151I3T1 A0A3B0KUT2 A0A195EE82 A0A154PAA7 A0A1J1IA90 B4ICG3 A0A195FWJ5
A0A0N1ID10 A0A194PXI9 A0A0N1PHB6 A0A2A4JVC9 A0A2W1BIG4 B3XXP3 A0A3S2NWT7 D6WNN7 A0A0S1YDI1 A0A023F8Q5 A0A1Y1LM32 A3RE79 A0A139WI74 T1HB42 A0A2J7QPH5 A3RE80 U3UA05 K9JB71 E2AMN8 S5XYH3 A0A1B6MMH0 A0A3L8DHW8 E2BEV4 A0A1I8NBB0 N1NSV9 A0A026VWL4 A0A182XBF7 A0A0P5DAL8 A0A182F845 A0A0K8WE96 A0A0A1XTA7 A0A0P5XV02 A0A182VYW8 A0A212EZY2 A0A0P5DKE7 A0A182V4S4 A0A0P5UCK7 A0A182I098 A0A0P5D3F6 A0A0P5VKY2 A0A0N8EQ24 B0WG36 A0A0P6BPA6 A0A034WA99 A0A182UA01 Q7PYI9 A0A182RPX1 A0A182Y5B5 A0A1I8P294 A0A1A9UN78 U3U4F2 A0A182PVL2 A0A067RFL8 A0A0P5S7T5 A0A1B0G6W9 Q1RKW4 A0A0R1E5S3 A0A0P5S165 A0A0P5D985 A0A182F844 A0A1B0BRH7 A0A1A9YK95 A0A182LU62 A0A182NS99 A0A0P5IEN0 B3LYJ1 A0A3B0K5U2 A0A182PZC5 A0A1B0AAA9 B4QVZ9 A0A182MEC2 A0A1A9W8A8 A0A182I097 B4K6B4 X1WN36 B5DX60 A0A182U3G5 A0A182XBF6 B4GEI1 B4NJA1 A0A2A3EK67 H5V8A1 A0A182KLW8 A0A182PVL3 F4WNW6 Q868T3 A0A151I3T1 A0A3B0KUT2 A0A195EE82 A0A154PAA7 A0A1J1IA90 B4ICG3 A0A195FWJ5
Pubmed
18725956
28756777
26354079
22118469
18362917
19820115
+ More
25474469 28004739 18054377 18025266 18316733 20068045 21843505 23604020 23932938 20798317 23874803 30249741 25315136 24508170 25830018 25348373 12364791 25244985 24845553 17994087 17550304 15632085 20966253 21719571 12177421 12646179 10731132 12537572
25474469 28004739 18054377 18025266 18316733 20068045 21843505 23604020 23932938 20798317 23874803 30249741 25315136 24508170 25830018 25348373 12364791 25244985 24845553 17994087 17550304 15632085 20966253 21719571 12177421 12646179 10731132 12537572
EMBL
AB330447
BAG68425.1
ODYU01002644
SOQ40421.1
KZ149981
PZC75801.1
+ More
KQ459586 KPI98043.1 KT031024 RSAL01000125 ALM88322.1 RVE46572.1 AGBW02011206 OWR47041.1 KQ460874 KPJ11677.1 KPI98042.1 KPJ11676.1 NWSH01000541 PCG75769.1 KZ150134 PZC73017.1 AB330451 BAG68429.1 RVE46570.1 GQ331027 KQ971342 ADH10240.1 EFA04640.2 KT031028 ALM88326.1 GBBI01000921 JAC17791.1 GEZM01051714 JAV74734.1 EF222291 BK006104 ABN79651.1 DAA64485.1 KYB27541.1 ACPB03000022 ACPB03000023 NEVH01012089 PNF30488.1 EF222292 ABN79652.1 AB817311 BAO01078.1 GQ368180 ADI48561.1 GL440840 EFN65306.1 KC004225 AGT02811.1 GEBQ01021042 GEBQ01002918 JAT18935.1 JAT37059.1 QOIP01000008 RLU19753.1 GL447881 EFN85758.1 BK006103 DAA64484.1 KK107672 EZA48173.1 GDIP01158691 JAJ64711.1 GDHF01031384 GDHF01002950 JAI20930.1 JAI49364.1 GBXI01012474 GBXI01001179 GBXI01000087 JAD01818.1 JAD13113.1 JAD14205.1 GDIP01068395 JAM35320.1 OWR47042.1 GDIP01158303 JAJ65099.1 GDIP01132242 JAL71472.1 APCN01002056 GDIP01163504 JAJ59898.1 GDIP01098590 JAM05125.1 GDIQ01005076 JAN89661.1 DS231922 EDS26677.1 GDIP01012238 JAM91477.1 GAKP01007329 GAKP01007328 JAC51624.1 AAAB01008987 EAA01145.4 AB817309 BAO01076.1 KK852716 KDR17833.1 GDIQ01095536 JAL56190.1 CCAG010013874 BT025096 ABE73267.1 CM000160 KRK04421.1 GDIQ01111230 JAL40496.1 GDIP01159194 JAJ64208.1 JXJN01019093 AXCM01007076 GDIQ01218453 JAK33272.1 CH902617 EDV42906.2 OUUW01000023 SPP89597.1 AXCN02000205 CM000364 EDX14498.1 AXCM01007077 CH933806 EDW15187.2 ABLF02034364 ABLF02034366 ABLF02034367 ABLF02034369 ABLF02034370 CM000070 EDY68220.1 CH479182 EDW34016.1 CH964272 EDW84932.1 KZ288222 PBC32098.1 BT133202 AFA28443.1 GL888239 EGI64124.1 AF522188 AY219842 AE014297 AAN10041.1 AAO66429.1 KQ976475 KYM83903.1 SPP89596.1 KQ979039 KYN23416.1 KQ434856 KZC08773.1 CVRI01000047 CRK97131.1 CH480828 EDW45059.1 KQ981208 KYN44801.1
KQ459586 KPI98043.1 KT031024 RSAL01000125 ALM88322.1 RVE46572.1 AGBW02011206 OWR47041.1 KQ460874 KPJ11677.1 KPI98042.1 KPJ11676.1 NWSH01000541 PCG75769.1 KZ150134 PZC73017.1 AB330451 BAG68429.1 RVE46570.1 GQ331027 KQ971342 ADH10240.1 EFA04640.2 KT031028 ALM88326.1 GBBI01000921 JAC17791.1 GEZM01051714 JAV74734.1 EF222291 BK006104 ABN79651.1 DAA64485.1 KYB27541.1 ACPB03000022 ACPB03000023 NEVH01012089 PNF30488.1 EF222292 ABN79652.1 AB817311 BAO01078.1 GQ368180 ADI48561.1 GL440840 EFN65306.1 KC004225 AGT02811.1 GEBQ01021042 GEBQ01002918 JAT18935.1 JAT37059.1 QOIP01000008 RLU19753.1 GL447881 EFN85758.1 BK006103 DAA64484.1 KK107672 EZA48173.1 GDIP01158691 JAJ64711.1 GDHF01031384 GDHF01002950 JAI20930.1 JAI49364.1 GBXI01012474 GBXI01001179 GBXI01000087 JAD01818.1 JAD13113.1 JAD14205.1 GDIP01068395 JAM35320.1 OWR47042.1 GDIP01158303 JAJ65099.1 GDIP01132242 JAL71472.1 APCN01002056 GDIP01163504 JAJ59898.1 GDIP01098590 JAM05125.1 GDIQ01005076 JAN89661.1 DS231922 EDS26677.1 GDIP01012238 JAM91477.1 GAKP01007329 GAKP01007328 JAC51624.1 AAAB01008987 EAA01145.4 AB817309 BAO01076.1 KK852716 KDR17833.1 GDIQ01095536 JAL56190.1 CCAG010013874 BT025096 ABE73267.1 CM000160 KRK04421.1 GDIQ01111230 JAL40496.1 GDIP01159194 JAJ64208.1 JXJN01019093 AXCM01007076 GDIQ01218453 JAK33272.1 CH902617 EDV42906.2 OUUW01000023 SPP89597.1 AXCN02000205 CM000364 EDX14498.1 AXCM01007077 CH933806 EDW15187.2 ABLF02034364 ABLF02034366 ABLF02034367 ABLF02034369 ABLF02034370 CM000070 EDY68220.1 CH479182 EDW34016.1 CH964272 EDW84932.1 KZ288222 PBC32098.1 BT133202 AFA28443.1 GL888239 EGI64124.1 AF522188 AY219842 AE014297 AAN10041.1 AAO66429.1 KQ976475 KYM83903.1 SPP89596.1 KQ979039 KYN23416.1 KQ434856 KZC08773.1 CVRI01000047 CRK97131.1 CH480828 EDW45059.1 KQ981208 KYN44801.1
Proteomes
UP000053268
UP000283053
UP000007151
UP000053240
UP000218220
UP000007266
+ More
UP000015103 UP000235965 UP000000311 UP000279307 UP000008237 UP000095301 UP000053097 UP000076407 UP000069272 UP000075920 UP000075903 UP000075840 UP000002320 UP000075902 UP000007062 UP000075900 UP000076408 UP000095300 UP000078200 UP000075885 UP000027135 UP000092444 UP000002282 UP000092460 UP000092443 UP000075883 UP000075884 UP000007801 UP000268350 UP000075886 UP000092445 UP000000304 UP000091820 UP000009192 UP000007819 UP000001819 UP000008744 UP000007798 UP000242457 UP000075882 UP000007755 UP000000803 UP000078540 UP000078492 UP000076502 UP000183832 UP000001292 UP000078541
UP000015103 UP000235965 UP000000311 UP000279307 UP000008237 UP000095301 UP000053097 UP000076407 UP000069272 UP000075920 UP000075903 UP000075840 UP000002320 UP000075902 UP000007062 UP000075900 UP000076408 UP000095300 UP000078200 UP000075885 UP000027135 UP000092444 UP000002282 UP000092460 UP000092443 UP000075883 UP000075884 UP000007801 UP000268350 UP000075886 UP000092445 UP000000304 UP000091820 UP000009192 UP000007819 UP000001819 UP000008744 UP000007798 UP000242457 UP000075882 UP000007755 UP000000803 UP000078540 UP000078492 UP000076502 UP000183832 UP000001292 UP000078541
Pfam
PF00001 7tm_1
ProteinModelPortal
B3XXN9
A0A2H1VI11
A0A2W1BNF2
A0A194PYE0
A0A0S1YDW4
A0A212F010
+ More
A0A0N1ID10 A0A194PXI9 A0A0N1PHB6 A0A2A4JVC9 A0A2W1BIG4 B3XXP3 A0A3S2NWT7 D6WNN7 A0A0S1YDI1 A0A023F8Q5 A0A1Y1LM32 A3RE79 A0A139WI74 T1HB42 A0A2J7QPH5 A3RE80 U3UA05 K9JB71 E2AMN8 S5XYH3 A0A1B6MMH0 A0A3L8DHW8 E2BEV4 A0A1I8NBB0 N1NSV9 A0A026VWL4 A0A182XBF7 A0A0P5DAL8 A0A182F845 A0A0K8WE96 A0A0A1XTA7 A0A0P5XV02 A0A182VYW8 A0A212EZY2 A0A0P5DKE7 A0A182V4S4 A0A0P5UCK7 A0A182I098 A0A0P5D3F6 A0A0P5VKY2 A0A0N8EQ24 B0WG36 A0A0P6BPA6 A0A034WA99 A0A182UA01 Q7PYI9 A0A182RPX1 A0A182Y5B5 A0A1I8P294 A0A1A9UN78 U3U4F2 A0A182PVL2 A0A067RFL8 A0A0P5S7T5 A0A1B0G6W9 Q1RKW4 A0A0R1E5S3 A0A0P5S165 A0A0P5D985 A0A182F844 A0A1B0BRH7 A0A1A9YK95 A0A182LU62 A0A182NS99 A0A0P5IEN0 B3LYJ1 A0A3B0K5U2 A0A182PZC5 A0A1B0AAA9 B4QVZ9 A0A182MEC2 A0A1A9W8A8 A0A182I097 B4K6B4 X1WN36 B5DX60 A0A182U3G5 A0A182XBF6 B4GEI1 B4NJA1 A0A2A3EK67 H5V8A1 A0A182KLW8 A0A182PVL3 F4WNW6 Q868T3 A0A151I3T1 A0A3B0KUT2 A0A195EE82 A0A154PAA7 A0A1J1IA90 B4ICG3 A0A195FWJ5
A0A0N1ID10 A0A194PXI9 A0A0N1PHB6 A0A2A4JVC9 A0A2W1BIG4 B3XXP3 A0A3S2NWT7 D6WNN7 A0A0S1YDI1 A0A023F8Q5 A0A1Y1LM32 A3RE79 A0A139WI74 T1HB42 A0A2J7QPH5 A3RE80 U3UA05 K9JB71 E2AMN8 S5XYH3 A0A1B6MMH0 A0A3L8DHW8 E2BEV4 A0A1I8NBB0 N1NSV9 A0A026VWL4 A0A182XBF7 A0A0P5DAL8 A0A182F845 A0A0K8WE96 A0A0A1XTA7 A0A0P5XV02 A0A182VYW8 A0A212EZY2 A0A0P5DKE7 A0A182V4S4 A0A0P5UCK7 A0A182I098 A0A0P5D3F6 A0A0P5VKY2 A0A0N8EQ24 B0WG36 A0A0P6BPA6 A0A034WA99 A0A182UA01 Q7PYI9 A0A182RPX1 A0A182Y5B5 A0A1I8P294 A0A1A9UN78 U3U4F2 A0A182PVL2 A0A067RFL8 A0A0P5S7T5 A0A1B0G6W9 Q1RKW4 A0A0R1E5S3 A0A0P5S165 A0A0P5D985 A0A182F844 A0A1B0BRH7 A0A1A9YK95 A0A182LU62 A0A182NS99 A0A0P5IEN0 B3LYJ1 A0A3B0K5U2 A0A182PZC5 A0A1B0AAA9 B4QVZ9 A0A182MEC2 A0A1A9W8A8 A0A182I097 B4K6B4 X1WN36 B5DX60 A0A182U3G5 A0A182XBF6 B4GEI1 B4NJA1 A0A2A3EK67 H5V8A1 A0A182KLW8 A0A182PVL3 F4WNW6 Q868T3 A0A151I3T1 A0A3B0KUT2 A0A195EE82 A0A154PAA7 A0A1J1IA90 B4ICG3 A0A195FWJ5
PDB
5DHH
E-value=2.08947e-15,
Score=200
Ontologies
GO
Topology
Subcellular location
Cell membrane
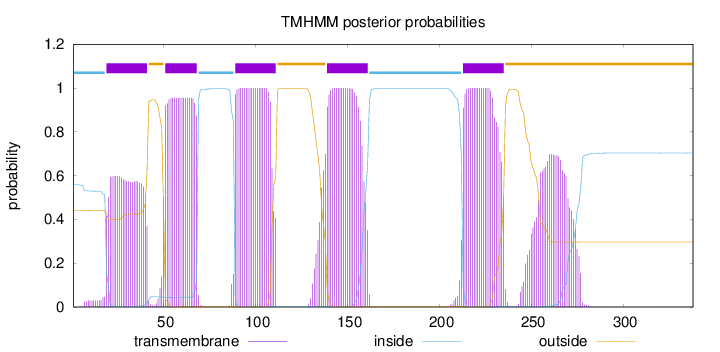
Length:
338
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
114.26395
Exp number, first 60 AAs:
23.53435
Total prob of N-in:
0.55820
POSSIBLE N-term signal
sequence
inside
1 - 18
TMhelix
19 - 41
outside
42 - 50
TMhelix
51 - 68
inside
69 - 88
TMhelix
89 - 111
outside
112 - 138
TMhelix
139 - 161
inside
162 - 212
TMhelix
213 - 235
outside
236 - 338
Population Genetic Test Statistics
Pi
21.826025
Theta
20.559421
Tajima's D
0.353016
CLR
0.62262
CSRT
0.476326183690815
Interpretation
Uncertain
