Gene
KWMTBOMO13435
Annotation
PREDICTED:_neuropeptide_receptor_A30_isoform_X1_[Bombyx_mori]
Full name
Cardioacceleratory peptide receptor
+ More
Alkaline phosphatase
Alkaline phosphatase
Alternative Name
Crustacean cardioactive peptide receptor
Location in the cell
PlasmaMembrane Reliability : 3.557
Sequence
CDS
ATGACATTTGTCATAGTCTTCGTATTCGTCCTCTGCTGGTCTCCCTACATGATCTTCGATCTACTTCAAGTATATGGCTACGTTCCGGACACACAAGTGAACGTGGCCATAGCGTCTCTCATACAAAGTCTCGCCCCGCTCAACTCGGCAGCGAATCCGGTCATCTACTTTATATTCTCGAACCGGATCTTCGTGAGTCTCAAGAACATCCCTCCATACAAGTGGCTGACTTGTCTGATGAAGGGTAATGATAGTCCGGCTGGCAATGAATCCAGGGCCCATACGGAGCTCCTCACGTCCTCACATCGAAGAACCAGGCGCGATCTTACCATAAGGATATGA
Protein
MTFVIVFVFVLCWSPYMIFDLLQVYGYVPDTQVNVAIASLIQSLAPLNSAANPVIYFIFSNRIFVSLKNIPPYKWLTCLMKGNDSPAGNESRAHTELLTSSHRRTRRDLTIRI
Summary
Description
Binds to the cardioactive peptide (CCAP), which is a neuropeptide.
Catalytic Activity
a phosphate monoester + H2O = an alcohol + phosphate
Similarity
Belongs to the G-protein coupled receptor 1 family. Vasopressin/oxytocin receptor subfamily.
Belongs to the alkaline phosphatase family.
Belongs to the alkaline phosphatase family.
Keywords
Cell membrane
Complete proteome
G-protein coupled receptor
Glycoprotein
Membrane
Receptor
Reference proteome
Transducer
Transmembrane
Transmembrane helix
Feature
chain Cardioacceleratory peptide receptor
Uniprot
B3XXP3
A0A3S2NWT7
A0A0S1YDI1
A0A2A4JVC9
A0A194PXI9
A0A2W1BIG4
+ More
A0A0N1PHB6 A0A212EZY2 A0A2H1VZF2 A0A2S2PL43 X1X8G0 X1WN36 A0A0A9WWH4 A0A2W1BNF2 A0A2H1VI11 A0A194PYE0 A0A0A9ZFP9 A0A0A9WWH9 A0A0N1ID10 A0A023F8Q5 B3XXN9 A0A2A4JWG3 S5XYH3 T1HB42 A0A1B6HSY4 A0A212F010 A0A1B6FJH5 B4K6B4 A0A0K8V2G7 A0A1B6LW69 B4LYQ8 N1NSV9 A0A0M4EF51 Q1RKW4 A0A0R1E5S3 A0A1I8NBB0 D6WNN7 A0A1I8P294 A0A1A9UN78 B4QVZ9 A3RE79 H5V8A1 Q868T3 A0A3B0K5U2 B4ICG3 A0A0S1YDW4 A0A1B0BRH7 B4NJA1 B5DX60 A0A1B0AAA9 B4PSZ2 A0A034WA99 B3LYJ1 A0A1A9YK95 A0A1B0G6W9 B4JGH4 B4GEI1 A0A3B0KUT2 A0A1W4V962 B3P6C8 A0A1B6MMH0 K9JB71 A0A0L0CPL6 A0A0K8WE96 A0A0A1XTA7 A0A1A9W8A8 N6U5R9 A0A139WDX0 Q16Y65 A0A1S4FK74 A0A1Y1LM32 A0A182NS99 A0A182PZC5 B0WG36 A0A182PVL3 A0A182MEC2 A0A182F844 A0A182KD15 A0A2R7WKR3 A0A182VYW9 A0A182I097 A0A139WI74 A0A182XBF6 A0A182U3G5 A0A182RPX0 A0A182KLW8 W5JBF8 A0A182Y5B6 A0A182VM83 A0NHA2 A0A182LU62 A0A182RPX1 Q1JQQ1 A0A182F845 Q7PYJ0 A0A182VYW8
A0A0N1PHB6 A0A212EZY2 A0A2H1VZF2 A0A2S2PL43 X1X8G0 X1WN36 A0A0A9WWH4 A0A2W1BNF2 A0A2H1VI11 A0A194PYE0 A0A0A9ZFP9 A0A0A9WWH9 A0A0N1ID10 A0A023F8Q5 B3XXN9 A0A2A4JWG3 S5XYH3 T1HB42 A0A1B6HSY4 A0A212F010 A0A1B6FJH5 B4K6B4 A0A0K8V2G7 A0A1B6LW69 B4LYQ8 N1NSV9 A0A0M4EF51 Q1RKW4 A0A0R1E5S3 A0A1I8NBB0 D6WNN7 A0A1I8P294 A0A1A9UN78 B4QVZ9 A3RE79 H5V8A1 Q868T3 A0A3B0K5U2 B4ICG3 A0A0S1YDW4 A0A1B0BRH7 B4NJA1 B5DX60 A0A1B0AAA9 B4PSZ2 A0A034WA99 B3LYJ1 A0A1A9YK95 A0A1B0G6W9 B4JGH4 B4GEI1 A0A3B0KUT2 A0A1W4V962 B3P6C8 A0A1B6MMH0 K9JB71 A0A0L0CPL6 A0A0K8WE96 A0A0A1XTA7 A0A1A9W8A8 N6U5R9 A0A139WDX0 Q16Y65 A0A1S4FK74 A0A1Y1LM32 A0A182NS99 A0A182PZC5 B0WG36 A0A182PVL3 A0A182MEC2 A0A182F844 A0A182KD15 A0A2R7WKR3 A0A182VYW9 A0A182I097 A0A139WI74 A0A182XBF6 A0A182U3G5 A0A182RPX0 A0A182KLW8 W5JBF8 A0A182Y5B6 A0A182VM83 A0NHA2 A0A182LU62 A0A182RPX1 Q1JQQ1 A0A182F845 Q7PYJ0 A0A182VYW8
EC Number
3.1.3.1
Pubmed
18725956
26354079
28756777
22118469
25401762
25474469
+ More
23874803 17994087 18054377 18025266 18316733 20068045 21843505 23604020 17550304 25315136 18362917 19820115 12177421 12646179 10731132 12537572 15632085 25348373 26108605 25830018 23537049 17510324 28004739 20966253 20920257 23761445 25244985 12364791 16616003 14747013 17210077
23874803 17994087 18054377 18025266 18316733 20068045 21843505 23604020 17550304 25315136 18362917 19820115 12177421 12646179 10731132 12537572 15632085 25348373 26108605 25830018 23537049 17510324 28004739 20966253 20920257 23761445 25244985 12364791 16616003 14747013 17210077
EMBL
AB330451
BAG68429.1
RSAL01000125
RVE46570.1
KT031028
ALM88326.1
+ More
NWSH01000541 PCG75769.1 KQ459586 KPI98042.1 KZ150134 PZC73017.1 KQ460874 KPJ11676.1 AGBW02011206 OWR47042.1 ODYU01005263 SOQ45942.1 GGMR01017562 MBY30181.1 ABLF02014736 ABLF02034364 ABLF02034366 ABLF02034367 ABLF02034369 ABLF02034370 GBHO01031818 JAG11786.1 KZ149981 PZC75801.1 ODYU01002644 SOQ40421.1 KPI98043.1 GBHO01001321 JAG42283.1 GBHO01031813 JAG11791.1 KPJ11677.1 GBBI01000921 JAC17791.1 AB330447 BAG68425.1 NWSH01000447 PCG76341.1 KC004225 AGT02811.1 ACPB03000022 ACPB03000023 GECU01029951 JAS77755.1 OWR47041.1 GECZ01019427 JAS50342.1 CH933806 EDW15187.2 GDHF01019273 JAI33041.1 GEBQ01012053 JAT27924.1 CH940650 EDW66985.1 BK006103 DAA64484.1 CP012526 ALC46754.1 BT025096 ABE73267.1 CM000160 KRK04421.1 GQ331027 KQ971342 ADH10240.1 EFA04640.2 CM000364 EDX14498.1 EF222291 BK006104 ABN79651.1 DAA64485.1 BT133202 AFA28443.1 AF522188 AY219842 AE014297 AAN10041.1 AAO66429.1 OUUW01000023 SPP89597.1 CH480828 EDW45059.1 KT031024 ALM88322.1 RVE46572.1 JXJN01019093 CH964272 EDW84932.1 CM000070 EDY68220.1 EDW98679.2 GAKP01007329 GAKP01007328 JAC51624.1 CH902617 EDV42906.2 CCAG010013874 CH916369 EDV93671.1 CH479182 EDW34016.1 SPP89596.1 CH954182 EDV53598.1 GEBQ01021042 GEBQ01002918 JAT18935.1 JAT37059.1 GQ368180 ADI48561.1 JRES01000091 KNC34201.1 GDHF01031384 GDHF01002950 JAI20930.1 JAI49364.1 GBXI01012474 GBXI01001179 GBXI01000087 JAD01818.1 JAD13113.1 JAD14205.1 APGK01041601 APGK01041602 KB740994 ENN75991.1 KQ971357 KYB26031.1 CH477524 EAT39545.2 GEZM01051714 JAV74734.1 AXCN02000205 DS231922 EDS26677.1 AXCM01007076 AXCM01007077 KK854897 PTY19580.1 APCN01002056 KYB27541.1 ADMH02001611 ETN61802.1 AAAB01008987 EAU75602.2 AY500851 AAS77205.1 EAA01142.4
NWSH01000541 PCG75769.1 KQ459586 KPI98042.1 KZ150134 PZC73017.1 KQ460874 KPJ11676.1 AGBW02011206 OWR47042.1 ODYU01005263 SOQ45942.1 GGMR01017562 MBY30181.1 ABLF02014736 ABLF02034364 ABLF02034366 ABLF02034367 ABLF02034369 ABLF02034370 GBHO01031818 JAG11786.1 KZ149981 PZC75801.1 ODYU01002644 SOQ40421.1 KPI98043.1 GBHO01001321 JAG42283.1 GBHO01031813 JAG11791.1 KPJ11677.1 GBBI01000921 JAC17791.1 AB330447 BAG68425.1 NWSH01000447 PCG76341.1 KC004225 AGT02811.1 ACPB03000022 ACPB03000023 GECU01029951 JAS77755.1 OWR47041.1 GECZ01019427 JAS50342.1 CH933806 EDW15187.2 GDHF01019273 JAI33041.1 GEBQ01012053 JAT27924.1 CH940650 EDW66985.1 BK006103 DAA64484.1 CP012526 ALC46754.1 BT025096 ABE73267.1 CM000160 KRK04421.1 GQ331027 KQ971342 ADH10240.1 EFA04640.2 CM000364 EDX14498.1 EF222291 BK006104 ABN79651.1 DAA64485.1 BT133202 AFA28443.1 AF522188 AY219842 AE014297 AAN10041.1 AAO66429.1 OUUW01000023 SPP89597.1 CH480828 EDW45059.1 KT031024 ALM88322.1 RVE46572.1 JXJN01019093 CH964272 EDW84932.1 CM000070 EDY68220.1 EDW98679.2 GAKP01007329 GAKP01007328 JAC51624.1 CH902617 EDV42906.2 CCAG010013874 CH916369 EDV93671.1 CH479182 EDW34016.1 SPP89596.1 CH954182 EDV53598.1 GEBQ01021042 GEBQ01002918 JAT18935.1 JAT37059.1 GQ368180 ADI48561.1 JRES01000091 KNC34201.1 GDHF01031384 GDHF01002950 JAI20930.1 JAI49364.1 GBXI01012474 GBXI01001179 GBXI01000087 JAD01818.1 JAD13113.1 JAD14205.1 APGK01041601 APGK01041602 KB740994 ENN75991.1 KQ971357 KYB26031.1 CH477524 EAT39545.2 GEZM01051714 JAV74734.1 AXCN02000205 DS231922 EDS26677.1 AXCM01007076 AXCM01007077 KK854897 PTY19580.1 APCN01002056 KYB27541.1 ADMH02001611 ETN61802.1 AAAB01008987 EAU75602.2 AY500851 AAS77205.1 EAA01142.4
Proteomes
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
UP000007819
+ More
UP000015103 UP000009192 UP000008792 UP000092553 UP000002282 UP000095301 UP000007266 UP000095300 UP000078200 UP000000304 UP000000803 UP000268350 UP000001292 UP000092460 UP000007798 UP000001819 UP000092445 UP000007801 UP000092443 UP000092444 UP000001070 UP000008744 UP000192221 UP000008711 UP000037069 UP000091820 UP000019118 UP000008820 UP000075884 UP000075886 UP000002320 UP000075885 UP000075883 UP000069272 UP000075881 UP000075920 UP000075840 UP000076407 UP000075902 UP000075900 UP000075882 UP000000673 UP000076408 UP000075903 UP000007062
UP000015103 UP000009192 UP000008792 UP000092553 UP000002282 UP000095301 UP000007266 UP000095300 UP000078200 UP000000304 UP000000803 UP000268350 UP000001292 UP000092460 UP000007798 UP000001819 UP000092445 UP000007801 UP000092443 UP000092444 UP000001070 UP000008744 UP000192221 UP000008711 UP000037069 UP000091820 UP000019118 UP000008820 UP000075884 UP000075886 UP000002320 UP000075885 UP000075883 UP000069272 UP000075881 UP000075920 UP000075840 UP000076407 UP000075902 UP000075900 UP000075882 UP000000673 UP000076408 UP000075903 UP000007062
Interpro
IPR017452
GPCR_Rhodpsn_7TM
+ More
IPR000276 GPCR_Rhodpsn
IPR001817 Vasoprsn_rcpt
IPR024088 Tyr-tRNA-ligase_bac-type
IPR018299 Alkaline_phosphatase_AS
IPR001952 Alkaline_phosphatase
IPR017850 Alkaline_phosphatase_core_sf
IPR002305 aa-tRNA-synth_Ic
IPR002307 Tyr-tRNA-ligase
IPR036986 S4_RNA-bd_sf
IPR014729 Rossmann-like_a/b/a_fold
IPR001412 aa-tRNA-synth_I_CS
IPR000276 GPCR_Rhodpsn
IPR001817 Vasoprsn_rcpt
IPR024088 Tyr-tRNA-ligase_bac-type
IPR018299 Alkaline_phosphatase_AS
IPR001952 Alkaline_phosphatase
IPR017850 Alkaline_phosphatase_core_sf
IPR002305 aa-tRNA-synth_Ic
IPR002307 Tyr-tRNA-ligase
IPR036986 S4_RNA-bd_sf
IPR014729 Rossmann-like_a/b/a_fold
IPR001412 aa-tRNA-synth_I_CS
SUPFAM
SSF53649
SSF53649
Gene 3D
ProteinModelPortal
B3XXP3
A0A3S2NWT7
A0A0S1YDI1
A0A2A4JVC9
A0A194PXI9
A0A2W1BIG4
+ More
A0A0N1PHB6 A0A212EZY2 A0A2H1VZF2 A0A2S2PL43 X1X8G0 X1WN36 A0A0A9WWH4 A0A2W1BNF2 A0A2H1VI11 A0A194PYE0 A0A0A9ZFP9 A0A0A9WWH9 A0A0N1ID10 A0A023F8Q5 B3XXN9 A0A2A4JWG3 S5XYH3 T1HB42 A0A1B6HSY4 A0A212F010 A0A1B6FJH5 B4K6B4 A0A0K8V2G7 A0A1B6LW69 B4LYQ8 N1NSV9 A0A0M4EF51 Q1RKW4 A0A0R1E5S3 A0A1I8NBB0 D6WNN7 A0A1I8P294 A0A1A9UN78 B4QVZ9 A3RE79 H5V8A1 Q868T3 A0A3B0K5U2 B4ICG3 A0A0S1YDW4 A0A1B0BRH7 B4NJA1 B5DX60 A0A1B0AAA9 B4PSZ2 A0A034WA99 B3LYJ1 A0A1A9YK95 A0A1B0G6W9 B4JGH4 B4GEI1 A0A3B0KUT2 A0A1W4V962 B3P6C8 A0A1B6MMH0 K9JB71 A0A0L0CPL6 A0A0K8WE96 A0A0A1XTA7 A0A1A9W8A8 N6U5R9 A0A139WDX0 Q16Y65 A0A1S4FK74 A0A1Y1LM32 A0A182NS99 A0A182PZC5 B0WG36 A0A182PVL3 A0A182MEC2 A0A182F844 A0A182KD15 A0A2R7WKR3 A0A182VYW9 A0A182I097 A0A139WI74 A0A182XBF6 A0A182U3G5 A0A182RPX0 A0A182KLW8 W5JBF8 A0A182Y5B6 A0A182VM83 A0NHA2 A0A182LU62 A0A182RPX1 Q1JQQ1 A0A182F845 Q7PYJ0 A0A182VYW8
A0A0N1PHB6 A0A212EZY2 A0A2H1VZF2 A0A2S2PL43 X1X8G0 X1WN36 A0A0A9WWH4 A0A2W1BNF2 A0A2H1VI11 A0A194PYE0 A0A0A9ZFP9 A0A0A9WWH9 A0A0N1ID10 A0A023F8Q5 B3XXN9 A0A2A4JWG3 S5XYH3 T1HB42 A0A1B6HSY4 A0A212F010 A0A1B6FJH5 B4K6B4 A0A0K8V2G7 A0A1B6LW69 B4LYQ8 N1NSV9 A0A0M4EF51 Q1RKW4 A0A0R1E5S3 A0A1I8NBB0 D6WNN7 A0A1I8P294 A0A1A9UN78 B4QVZ9 A3RE79 H5V8A1 Q868T3 A0A3B0K5U2 B4ICG3 A0A0S1YDW4 A0A1B0BRH7 B4NJA1 B5DX60 A0A1B0AAA9 B4PSZ2 A0A034WA99 B3LYJ1 A0A1A9YK95 A0A1B0G6W9 B4JGH4 B4GEI1 A0A3B0KUT2 A0A1W4V962 B3P6C8 A0A1B6MMH0 K9JB71 A0A0L0CPL6 A0A0K8WE96 A0A0A1XTA7 A0A1A9W8A8 N6U5R9 A0A139WDX0 Q16Y65 A0A1S4FK74 A0A1Y1LM32 A0A182NS99 A0A182PZC5 B0WG36 A0A182PVL3 A0A182MEC2 A0A182F844 A0A182KD15 A0A2R7WKR3 A0A182VYW9 A0A182I097 A0A139WI74 A0A182XBF6 A0A182U3G5 A0A182RPX0 A0A182KLW8 W5JBF8 A0A182Y5B6 A0A182VM83 A0NHA2 A0A182LU62 A0A182RPX1 Q1JQQ1 A0A182F845 Q7PYJ0 A0A182VYW8
PDB
5WIV
E-value=0.000121438,
Score=101
Ontologies
GO
PANTHER
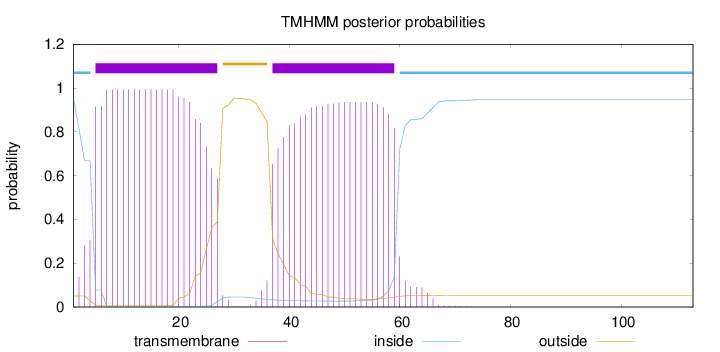
Topology
Subcellular location
Cell membrane
Length:
113
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.30752
Exp number, first 60 AAs:
42.77805
Total prob of N-in:
0.94929
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 36
TMhelix
37 - 59
inside
60 - 113
Population Genetic Test Statistics
Pi
24.945378
Theta
23.90727
Tajima's D
0.372191
CLR
1.679105
CSRT
0.48167591620419
Interpretation
Uncertain
