Gene
KWMTBOMO13416
Pre Gene Modal
BGIBMGA010901
Annotation
PREDICTED:_netrin_receptor_UNC5C-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.209
Sequence
CDS
ATGGAAGTGAACTTTTGGGTTTGCCTTTTAGTAGTTCATACTCAATTTTCGTTTTATGGGAAAGGACAGAAATTGAAAAATCAAAATCATCACGACGCAAAAACAGATGATCAAGATGAATTATCACCGGTTCATCATCAACCCCATATTAAGGATTACTTCCTGCCTCCGGTCTTCAATCACGATGACGAGGAGGAAGACGAAGATGAAGGTTACGCTGATTACGATTACGATAAAGGAACGAACTACAAGGATTCCCCTGATTTAGTTACAGTTCCCAGTGACACACAAGAAACCGAAGATAAAATACCATTACACACGACGAAAGACAATCTACCTGTATTTCTTTTAGAGCCGGAGAATACGTATGTGGTTAAAAATAAACCCGCTATTTTGAAATGTCGCGCTGCTAATGCTTTGGAGGTATACTTTAAATGCAATGGAATCAAAACACAAGGTGCGAACTTTGAATTCGTTGACCCACAAACCGGGGTGCGGATAATTGAAGGTGAACAATCGGTAACTAGAGAGAACATTGAAGAATACTTTGGTGGTGATAAGTACCAATGTTCCTGCTTCGCTTGGACCAGTCGAGGACAGATTAGAAGTCAGCCTGCTACAATTGAAATCGCATACATAAAGAAGCAATTCGCTGTAGCGCCGCAATCGCAACTGGTCAAACAGTACACATCAGTCAATTTCCGATGCGAGCCTCCACCAGCGGCGCCCTTCGCCCAGCTGTACTGGTTGAAGAATGGTGCTCCCGTTGTTATTGATGATAACGTTCAGGTTTCAACCGACGGAAATCTTATCATTAAGCAAGCTAATCTGCTGGACATGGCTAACTACACATGTGTTGCGGAAAACTTAGCCGGGAAGCGCATGTCAGAACCGGCTCTGCTGTCAGTATTCGTTAACGGTGGTTGGAGTACATGGTCAGCGTGGGTGGACTGTTGTCCAACAAGAGAAGGTCGAACGGGCAGAAAGAGAGAGAGAACTTGCACGGCACCGCGACCTCTAAACGGTGGACAGCCTTGCGTTGGCAACAATCTACAGAAGACTCAGGATTGTGTGGAGTGTGACATAGATTTGTATCTCGATAATATCGACGAGCTGGCTGATGAATCACCGAACATAGTTTTGGGACGTTGGTCGCAATGGTCAGACTGGTCGCGATGTGATTCCGACTGCCTTCAAACTAGAAGACGAAAGTGTCTCACGAGCTCGTGTCTGGGGAAAGACTACCAAGTCGCCCAGTGTCCTCTCTGTGTGCGCACGTCGAAATCTGAAAGAATTAATGACGGTACTCCGTATTGGCCCCTTTTCGTAGCGTTATCAATAGCCTCCCTCGTGTTTATAGTCGTAGTAATTCTCGGAATAAAATACATGAAGTTCAAGACGACTGAGACATCGCCTTACGCTAAACCGCCCACAGGTGGCAATTATTTCGGAAACGTTATAAAAAGGACTTTGACTAATCAACCAGACCTCACTATTCACGAGGAGTTCCACACGATAGATTCTTCACGGCTCAGGCACGCTAACATCATTACTAACGCACGGCATGAGCATTTGTACGAAGTGCCACAGCTAGCTAATAGTTACATAGCACCCATAGACCACGAGGTAAGGAACGACCGGATCGCTTGCAACAAGGACGCCTCTGAGTCAGACCGCTCTGACTCAAGCTGCTTTTTGAGCTCCGGTTCATCGTACAGCCACAGCGTAGAAATATCGCCTTCATTAAAAAACAATGCATCGTTAAACTCGAACTACGCAACGAATCTAGATACTTCCAAATCCAAGATGGTGTCGAGCGACGGCGACTGGTTAAATCTGGATAAAACCGGTGTTAGTCTGTTTGTTCCGGACGGCGTTGTTGATAAAGGAGAGGAATTGTTTTCTGTTGAAGTATCTGACGAGGAATGGAATAGGCCCATGCTTCAAGACGGCGAAATCCAATTGAGTCCGGTCATCAAATGCAGTCCTAAGAACTTTCACTTCCGTAAGGGCGTGGTCCTAACGTTTCCTCACTACGCATCGTTGAGGAACACCAGCTGGCTGCTATTCATCCTGCAGCGCCCTGATGATCTCCATACGAGGGAGTGGCGTAAAGTTTTGACTCTAGGCCAAGAGACTCCCGGCACGCCAATCTTCAGTCAAGTCGATCCTAACAAAGTGTACATCGTGGGCGAGTTCTTGAGCGATTTCGTCCTGGTGGGAAGAAGTTTTGCCGGTTCAGACGCCAAGAACCTAAAACTTGCCTTATTCATCTACGAAATGGACGAGAACTATCATCGTCTGAAACTGCACGTCTTCAACGATACGCCTCACTGTTTGAACGAGTGCTTCAATCTAGAACGGAGATTGGGCAGTAAGCTTCTGTGCGAACCGAAACCCTTCTATTTCCAGGAAAGCGGCTCAGATCTGAATAACGATGGTGGTATGTTGGTCGATCTTTCTCGTATATTCAGGGAGATGGGACGAATCGACTGTGCTACAGTTGTCGAGCGTAGATGCTTCTCGAATTAA
Protein
MEVNFWVCLLVVHTQFSFYGKGQKLKNQNHHDAKTDDQDELSPVHHQPHIKDYFLPPVFNHDDEEEDEDEGYADYDYDKGTNYKDSPDLVTVPSDTQETEDKIPLHTTKDNLPVFLLEPENTYVVKNKPAILKCRAANALEVYFKCNGIKTQGANFEFVDPQTGVRIIEGEQSVTRENIEEYFGGDKYQCSCFAWTSRGQIRSQPATIEIAYIKKQFAVAPQSQLVKQYTSVNFRCEPPPAAPFAQLYWLKNGAPVVIDDNVQVSTDGNLIIKQANLLDMANYTCVAENLAGKRMSEPALLSVFVNGGWSTWSAWVDCCPTREGRTGRKRERTCTAPRPLNGGQPCVGNNLQKTQDCVECDIDLYLDNIDELADESPNIVLGRWSQWSDWSRCDSDCLQTRRRKCLTSSCLGKDYQVAQCPLCVRTSKSERINDGTPYWPLFVALSIASLVFIVVVILGIKYMKFKTTETSPYAKPPTGGNYFGNVIKRTLTNQPDLTIHEEFHTIDSSRLRHANIITNARHEHLYEVPQLANSYIAPIDHEVRNDRIACNKDASESDRSDSSCFLSSGSSYSHSVEISPSLKNNASLNSNYATNLDTSKSKMVSSDGDWLNLDKTGVSLFVPDGVVDKGEELFSVEVSDEEWNRPMLQDGEIQLSPVIKCSPKNFHFRKGVVLTFPHYASLRNTSWLLFILQRPDDLHTREWRKVLTLGQETPGTPIFSQVDPNKVYIVGEFLSDFVLVGRSFAGSDAKNLKLALFIYEMDENYHRLKLHVFNDTPHCLNECFNLERRLGSKLLCEPKPFYFQESGSDLNNDGGMLVDLSRIFREMGRIDCATVVERRCFSN
Summary
Similarity
Belongs to the serpin family.
Uniprot
A0A2A4IZN1
A0A2W1BMT9
A0A194PXF9
A0A0N1I685
A0A3S2NWX0
A0A212EZX5
+ More
A0A212EVY5 A0A2W1BXA1 A0A2A4JM37 A0A2A4JLB3 A0A194Q8C5 A0A1Y1KGW7 A0A1B6LH58 A0A1B6KVK9 A0A195CJV7 A0A154PEH5 J9KU90 F4W406 A0A088A971 A0A151X1U2 A0A0L7RHT7 A0A158NTE6 A0A195DVW8 A0A195B423 A0A3L8DS02 W8AYM5 W8B2P2 A0A026VX76 A0A034W4Y3 U5EYZ9 A0A0K8UWV4 A0A0A1X6V4 A0A0P5B8K0 B4KS96 V3ZRF4 B4MFS3 A0A0Q9XFX8 Q28XS1 A0A3B0JI73 A0A0L0CDT6 B4GHH5 A0A026VUN0 A0A0R3NPS2 F4W410 B4MJ18 A0A195DVL0 A0A195B393 A0A3L8DRW3 A0A195EUI3 E2APQ4 A0A158NK16 A0A3Q3E6F9 H2LLZ0 H2LLY9 A0A3B5RDJ2 A0A1A7ZTP1 A0A3B3H6L5 A0A3Q1CPT3 A0A151X1E9 A0A3P8RPQ9 A0A195CJV1 A0A3B3YP05 A0A3B3VM99 A0A3B3XAI2 A0A3P9Q5P4 A0A3P9J0H9 A0A3Q1IQU9 A0A1A8C1H1 A0A1A8V5D7 A0A1A8Q8H3 A0A3P9Q5U0 A0A146ZII3 A0A3Q1IEH2 A0A3Q3WWQ5 A0A182NAQ8 A0A3Q3JB82 A0A3B4YRF8 A0A3B4VIA3 G3NYB5 A0A3Q3MXJ1 A0A3Q1IIL7 A0A3Q3JBA1 A0A3Q1FMK5 A0A3Q1C6E3 A0A154PEA8 A0A3Q0R8V6 A0A3Q4H2S8 A0A3Q2WVM9 A0A0L7QTL0
A0A212EVY5 A0A2W1BXA1 A0A2A4JM37 A0A2A4JLB3 A0A194Q8C5 A0A1Y1KGW7 A0A1B6LH58 A0A1B6KVK9 A0A195CJV7 A0A154PEH5 J9KU90 F4W406 A0A088A971 A0A151X1U2 A0A0L7RHT7 A0A158NTE6 A0A195DVW8 A0A195B423 A0A3L8DS02 W8AYM5 W8B2P2 A0A026VX76 A0A034W4Y3 U5EYZ9 A0A0K8UWV4 A0A0A1X6V4 A0A0P5B8K0 B4KS96 V3ZRF4 B4MFS3 A0A0Q9XFX8 Q28XS1 A0A3B0JI73 A0A0L0CDT6 B4GHH5 A0A026VUN0 A0A0R3NPS2 F4W410 B4MJ18 A0A195DVL0 A0A195B393 A0A3L8DRW3 A0A195EUI3 E2APQ4 A0A158NK16 A0A3Q3E6F9 H2LLZ0 H2LLY9 A0A3B5RDJ2 A0A1A7ZTP1 A0A3B3H6L5 A0A3Q1CPT3 A0A151X1E9 A0A3P8RPQ9 A0A195CJV1 A0A3B3YP05 A0A3B3VM99 A0A3B3XAI2 A0A3P9Q5P4 A0A3P9J0H9 A0A3Q1IQU9 A0A1A8C1H1 A0A1A8V5D7 A0A1A8Q8H3 A0A3P9Q5U0 A0A146ZII3 A0A3Q1IEH2 A0A3Q3WWQ5 A0A182NAQ8 A0A3Q3JB82 A0A3B4YRF8 A0A3B4VIA3 G3NYB5 A0A3Q3MXJ1 A0A3Q1IIL7 A0A3Q3JBA1 A0A3Q1FMK5 A0A3Q1C6E3 A0A154PEA8 A0A3Q0R8V6 A0A3Q4H2S8 A0A3Q2WVM9 A0A0L7QTL0
Pubmed
EMBL
NWSH01004342
PCG65211.1
KZ150134
PZC73003.1
KQ459586
KPI98031.1
+ More
KQ460874 KPJ11666.1 RSAL01000124 RVE46623.1 AGBW02011206 OWR47055.1 AGBW02012107 OWR45646.1 KZ149907 PZC78254.1 NWSH01001062 PCG72846.1 PCG72847.1 KQ459324 KPJ01644.1 GEZM01086422 JAV59460.1 GEBQ01016935 JAT23042.1 GEBQ01024488 JAT15489.1 KQ977642 KYN01011.1 KQ434870 KZC09610.1 ABLF02041018 GL887491 EGI71002.1 KQ982585 KYQ54236.1 KQ414585 KOC70552.1 ADTU01025884 ADTU01025885 KQ980295 KYN16887.1 KQ976625 KYM78959.1 QOIP01000005 RLU23184.1 GAMC01015248 JAB91307.1 GAMC01015247 JAB91308.1 KK107847 EZA47459.1 GAKP01009178 GAKP01009177 GAKP01009176 JAC49774.1 GANO01001690 JAB58181.1 GDHF01025673 GDHF01021499 JAI26641.1 JAI30815.1 GBXI01007470 JAD06822.1 GDIP01188765 JAJ34637.1 CH933808 EDW08378.1 KB203683 ESO83456.1 CH940667 EDW57244.1 KRG04084.1 CM000071 EAL26245.2 OUUW01000001 SPP75100.1 JRES01000644 KNC29659.1 CH479183 EDW35945.1 EZA47457.1 KRT02937.1 EGI71006.1 CH963719 EDW72107.1 KYN16881.1 KYM78963.1 RLU23185.1 KQ981979 KYN31539.1 GL441572 EFN64605.1 ADTU01018638 ADTU01018639 HADY01007659 SBP46144.1 KYQ54241.1 KYN01006.1 HADZ01008765 SBP72706.1 HAEJ01014574 SBS55031.1 HAEH01003003 HAEI01004676 SBR89826.1 GCES01020152 JAR66171.1 KZC09608.1 KQ414755 KOC61811.1
KQ460874 KPJ11666.1 RSAL01000124 RVE46623.1 AGBW02011206 OWR47055.1 AGBW02012107 OWR45646.1 KZ149907 PZC78254.1 NWSH01001062 PCG72846.1 PCG72847.1 KQ459324 KPJ01644.1 GEZM01086422 JAV59460.1 GEBQ01016935 JAT23042.1 GEBQ01024488 JAT15489.1 KQ977642 KYN01011.1 KQ434870 KZC09610.1 ABLF02041018 GL887491 EGI71002.1 KQ982585 KYQ54236.1 KQ414585 KOC70552.1 ADTU01025884 ADTU01025885 KQ980295 KYN16887.1 KQ976625 KYM78959.1 QOIP01000005 RLU23184.1 GAMC01015248 JAB91307.1 GAMC01015247 JAB91308.1 KK107847 EZA47459.1 GAKP01009178 GAKP01009177 GAKP01009176 JAC49774.1 GANO01001690 JAB58181.1 GDHF01025673 GDHF01021499 JAI26641.1 JAI30815.1 GBXI01007470 JAD06822.1 GDIP01188765 JAJ34637.1 CH933808 EDW08378.1 KB203683 ESO83456.1 CH940667 EDW57244.1 KRG04084.1 CM000071 EAL26245.2 OUUW01000001 SPP75100.1 JRES01000644 KNC29659.1 CH479183 EDW35945.1 EZA47457.1 KRT02937.1 EGI71006.1 CH963719 EDW72107.1 KYN16881.1 KYM78963.1 RLU23185.1 KQ981979 KYN31539.1 GL441572 EFN64605.1 ADTU01018638 ADTU01018639 HADY01007659 SBP46144.1 KYQ54241.1 KYN01006.1 HADZ01008765 SBP72706.1 HAEJ01014574 SBS55031.1 HAEH01003003 HAEI01004676 SBR89826.1 GCES01020152 JAR66171.1 KZC09608.1 KQ414755 KOC61811.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
UP000078542
+ More
UP000076502 UP000007819 UP000007755 UP000005203 UP000075809 UP000053825 UP000005205 UP000078492 UP000078540 UP000279307 UP000053097 UP000009192 UP000030746 UP000008792 UP000001819 UP000268350 UP000037069 UP000008744 UP000007798 UP000078541 UP000000311 UP000264820 UP000001038 UP000002852 UP000257160 UP000265080 UP000261480 UP000261500 UP000242638 UP000265200 UP000265040 UP000261620 UP000075884 UP000261600 UP000261360 UP000261420 UP000007635 UP000261640 UP000257200 UP000261340 UP000261580 UP000264840
UP000076502 UP000007819 UP000007755 UP000005203 UP000075809 UP000053825 UP000005205 UP000078492 UP000078540 UP000279307 UP000053097 UP000009192 UP000030746 UP000008792 UP000001819 UP000268350 UP000037069 UP000008744 UP000007798 UP000078541 UP000000311 UP000264820 UP000001038 UP000002852 UP000257160 UP000265080 UP000261480 UP000261500 UP000242638 UP000265200 UP000265040 UP000261620 UP000075884 UP000261600 UP000261360 UP000261420 UP000007635 UP000261640 UP000257200 UP000261340 UP000261580 UP000264840
Interpro
IPR000488
Death_domain
+ More
IPR007110 Ig-like_dom
IPR003598 Ig_sub2
IPR000906 ZU5_dom
IPR036179 Ig-like_dom_sf
IPR036383 TSP1_rpt_sf
IPR033772 UPA
IPR013098 Ig_I-set
IPR003599 Ig_sub
IPR013783 Ig-like_fold
IPR011029 DEATH-like_dom_sf
IPR000884 TSP1_rpt
IPR037936 UNC5
IPR042185 Serpin_sf_2
IPR042178 Serpin_sf_1
IPR023796 Serpin_dom
IPR036186 Serpin_sf
IPR007110 Ig-like_dom
IPR003598 Ig_sub2
IPR000906 ZU5_dom
IPR036179 Ig-like_dom_sf
IPR036383 TSP1_rpt_sf
IPR033772 UPA
IPR013098 Ig_I-set
IPR003599 Ig_sub
IPR013783 Ig-like_fold
IPR011029 DEATH-like_dom_sf
IPR000884 TSP1_rpt
IPR037936 UNC5
IPR042185 Serpin_sf_2
IPR042178 Serpin_sf_1
IPR023796 Serpin_dom
IPR036186 Serpin_sf
Gene 3D
ProteinModelPortal
A0A2A4IZN1
A0A2W1BMT9
A0A194PXF9
A0A0N1I685
A0A3S2NWX0
A0A212EZX5
+ More
A0A212EVY5 A0A2W1BXA1 A0A2A4JM37 A0A2A4JLB3 A0A194Q8C5 A0A1Y1KGW7 A0A1B6LH58 A0A1B6KVK9 A0A195CJV7 A0A154PEH5 J9KU90 F4W406 A0A088A971 A0A151X1U2 A0A0L7RHT7 A0A158NTE6 A0A195DVW8 A0A195B423 A0A3L8DS02 W8AYM5 W8B2P2 A0A026VX76 A0A034W4Y3 U5EYZ9 A0A0K8UWV4 A0A0A1X6V4 A0A0P5B8K0 B4KS96 V3ZRF4 B4MFS3 A0A0Q9XFX8 Q28XS1 A0A3B0JI73 A0A0L0CDT6 B4GHH5 A0A026VUN0 A0A0R3NPS2 F4W410 B4MJ18 A0A195DVL0 A0A195B393 A0A3L8DRW3 A0A195EUI3 E2APQ4 A0A158NK16 A0A3Q3E6F9 H2LLZ0 H2LLY9 A0A3B5RDJ2 A0A1A7ZTP1 A0A3B3H6L5 A0A3Q1CPT3 A0A151X1E9 A0A3P8RPQ9 A0A195CJV1 A0A3B3YP05 A0A3B3VM99 A0A3B3XAI2 A0A3P9Q5P4 A0A3P9J0H9 A0A3Q1IQU9 A0A1A8C1H1 A0A1A8V5D7 A0A1A8Q8H3 A0A3P9Q5U0 A0A146ZII3 A0A3Q1IEH2 A0A3Q3WWQ5 A0A182NAQ8 A0A3Q3JB82 A0A3B4YRF8 A0A3B4VIA3 G3NYB5 A0A3Q3MXJ1 A0A3Q1IIL7 A0A3Q3JBA1 A0A3Q1FMK5 A0A3Q1C6E3 A0A154PEA8 A0A3Q0R8V6 A0A3Q4H2S8 A0A3Q2WVM9 A0A0L7QTL0
A0A212EVY5 A0A2W1BXA1 A0A2A4JM37 A0A2A4JLB3 A0A194Q8C5 A0A1Y1KGW7 A0A1B6LH58 A0A1B6KVK9 A0A195CJV7 A0A154PEH5 J9KU90 F4W406 A0A088A971 A0A151X1U2 A0A0L7RHT7 A0A158NTE6 A0A195DVW8 A0A195B423 A0A3L8DS02 W8AYM5 W8B2P2 A0A026VX76 A0A034W4Y3 U5EYZ9 A0A0K8UWV4 A0A0A1X6V4 A0A0P5B8K0 B4KS96 V3ZRF4 B4MFS3 A0A0Q9XFX8 Q28XS1 A0A3B0JI73 A0A0L0CDT6 B4GHH5 A0A026VUN0 A0A0R3NPS2 F4W410 B4MJ18 A0A195DVL0 A0A195B393 A0A3L8DRW3 A0A195EUI3 E2APQ4 A0A158NK16 A0A3Q3E6F9 H2LLZ0 H2LLY9 A0A3B5RDJ2 A0A1A7ZTP1 A0A3B3H6L5 A0A3Q1CPT3 A0A151X1E9 A0A3P8RPQ9 A0A195CJV1 A0A3B3YP05 A0A3B3VM99 A0A3B3XAI2 A0A3P9Q5P4 A0A3P9J0H9 A0A3Q1IQU9 A0A1A8C1H1 A0A1A8V5D7 A0A1A8Q8H3 A0A3P9Q5U0 A0A146ZII3 A0A3Q1IEH2 A0A3Q3WWQ5 A0A182NAQ8 A0A3Q3JB82 A0A3B4YRF8 A0A3B4VIA3 G3NYB5 A0A3Q3MXJ1 A0A3Q1IIL7 A0A3Q3JBA1 A0A3Q1FMK5 A0A3Q1C6E3 A0A154PEA8 A0A3Q0R8V6 A0A3Q4H2S8 A0A3Q2WVM9 A0A0L7QTL0
PDB
5FTT
E-value=1.8314e-48,
Score=489
Ontologies
GO
PANTHER
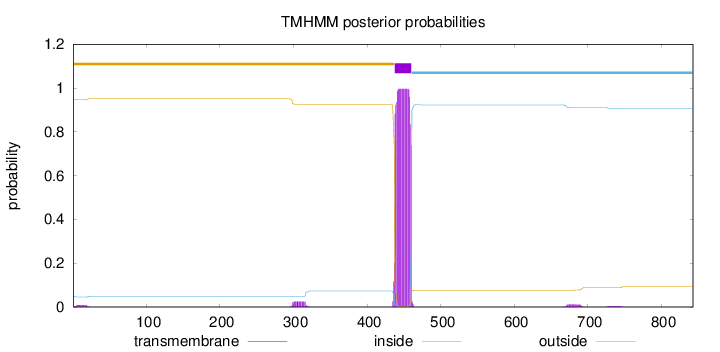
Topology
Length:
843
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.5622399999999
Exp number, first 60 AAs:
0.1434
Total prob of N-in:
0.05076
outside
1 - 437
TMhelix
438 - 460
inside
461 - 843
Population Genetic Test Statistics
Pi
23.852279
Theta
24.995141
Tajima's D
0.082378
CLR
0.567192
CSRT
0.39838008099595
Interpretation
Uncertain
