Gene
KWMTBOMO13415
Annotation
PREDICTED:_antigen_43-like_isoform_X2_[Salmo_salar]
Location in the cell
Extracellular Reliability : 1.848 Nuclear Reliability : 1.57
Sequence
CDS
ATGCTGCAAAGTCGCTCTGCAACGTCATGCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATGCTGCAACGTCATTCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATTCTGCAATGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCATGCTGCAACGTCATGCTGCAACGTCGCTCTGCAACATCATTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCATTCTGCAACGTCATTCTGCAACGTCATGCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCATGCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCATCGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCATTCTGCAACGTCATTCTGCAACGTCATGCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCATGCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCATCGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATGCTGCAACGTCATTCTGCACCGTCGCTCTGCAACGTCATGCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATGCTGCAACGTCATTCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATTCTGCAATGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCATGCTGCAACGTCATGCTGCAACGTCGCTCTGCAACATCATTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCATTCTGCAACGTCATTCTGCAACGTCATGCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCATGCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATGCTGCATCGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCGCTATGCAACGTCATGCTGCAAAGTCGCTCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCGCTCTGCAACGTCGCTCTGCAACGTCATGCTGCAACGTCATTCTGCAACGTCATGCTGCAACGTCGCTATGCAACGTCATTCTGCAACGTCGCTCTGCAACGTCATGA
Protein
MLQSRSATSCCNVILQRRSATSCCNVAMQRHSATSLCNVMLQRRSATSFCNVALQRRYATSCCKVALQRHSATSLCNVILQRRSATSLCNVMLQSRSATSCCNVILQRRYATSFCNVALQRRYATSFCNVALQRHSATSLCNVILQCHSATSLCNVAMQRHAAKSLCNVAMQRHAAKSLCNVILQRRSATSCCNVALQRHSATSLCNVAMQRHAAKSLCNVILQRRSATSFCNVALQRRYATSCCKVALQRHSATSCCNVMLQRRSATSFCNVILQRRSATSFCNVALQRHAAKSLCNVILQRRSATSLCNVMLQRRYATSFCNVALQRHSATSFCNVALQRHAATSFCNVILQRHAATSLCNVILQRRSATSCCNVALQRHAATSLCNVMLQRRSATSCCNVALQRHAATSLCNVAMQRHAAKSLCNVILQRHAATSLCNVILQRHAATSLCNVMLQRRSATSCCNVALQRHAASSLCNVMLQRRSATSLCNVMLQRRYATSFCNVALQRHSATSFCNVILQRHAATSLCNVILQRRSATSCCNVALQRHAATSLCNVMLQRRSATSCCNVALQRHAATSLCNVAMQRHAAKSLCNVILQRHAATSLCNVILQRHAATSLCNVMLQRRSATSCCNVALQRHAASSLCNVMLQRRSATSFCNVALQRRYATSCCKVALQRHAATSFCTVALQRHAATSLCNVILQRRSATSCCNVALQRHSATSLCNVAMQRHAAKSLCNVILQRRSATSFCNVALQRRYATSCCKVALQRHAATSFCNVAMQRHSATSLCNVAMQRHSATSLCNVILQRRSATSFCNVILQRRSATSLCNVMLQSRSATSLCNVMLQSRSATSFCNVALQRHAATSLCNVILQRRSATSLCNVMLQSRSATSFCNVALQRHSATSLCNVAMQRHAAKSLCNVILQRHAATSCCNVALQHHSATSFCNVALQRHSATSLCNVMLQSRSATSFCNVALQRRYATSCCNVAMQRHSATSLCNVILQRHSATSLCNVMLQRHSATSFCNVMLQRRYATSFCNVALQRHAATSLCNVMLQRRSATSCCNVALQRHAATSLCNVMLQRRSATSLCNVMLQSRSATSFCNVMLQRRYATSFCNVMLQRRSATSCCNVALQRHAATSLCNVMLHRRSATSCCNVALQRHSATSLCNVAMQRHAAKSLCNVILQRRSATSCCNVILQRRSATSCCNVALQRRSATSCCNVILQRHAATSLCNVILQRRSATS
Summary
Catalytic Activity
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
EMBL
Proteomes
Interpro
IPR000242
PTPase_domain
+ More
IPR000387 TYR_PHOSPHATASE_dom
IPR016130 Tyr_Pase_AS
IPR003595 Tyr_Pase_cat
IPR029021 Prot-tyrosine_phosphatase-like
IPR011004 Trimer_LpxA-like_sf
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR000387 TYR_PHOSPHATASE_dom
IPR016130 Tyr_Pase_AS
IPR003595 Tyr_Pase_cat
IPR029021 Prot-tyrosine_phosphatase-like
IPR011004 Trimer_LpxA-like_sf
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
Gene 3D
ProteinModelPortal
Ontologies
PANTHER
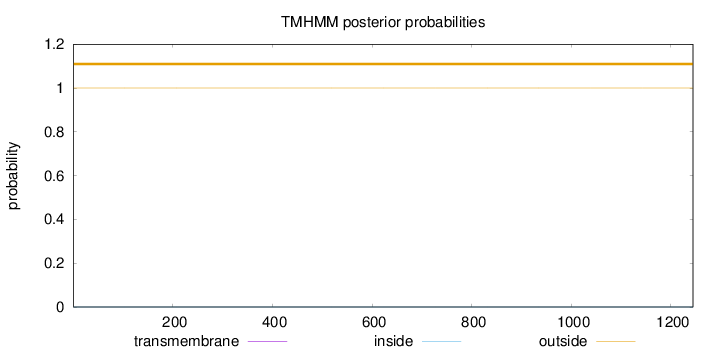
Topology
Length:
1244
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00255
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.00009
outside
1 - 1244
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
308.914345
CSRT
0
Interpretation
Uncertain
