Gene
KWMTBOMO13413
Pre Gene Modal
BGIBMGA010855
Annotation
Dual_specificity_protein_phosphatase_18_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.859
Sequence
CDS
ATGAAAGTTTTAACTGTTGGAGACAAAGTTGCTAGTGTCGGTCGAGAGAATGAGCCCACCGTTGTGGTCGACACGCAAAAGTTAATTAGAGAAGAATTGAACAGGGGCTGTCCATTGGGCGTGTCGAGGGTCACGAATCTAGTCTACGTGTGTGGAGCTCACGCTCTACCTGGAGCCGTCAGGGTTTTGCATCCTGGATTGATCGTCAGCGCGGCGCCAGAACTGCCTCCACCGCCCGAAGATTACGTACCGAGGCATTTCGTACCCCTGTTGGACACTCCGAACTCAGATATGCTCCCCTATATGGATAGGGTTGCTGACCTTATTAATGAGGTGGTCACAAACGGAGGGGTGGTGTTGGTTCATTGCGTCGCCGGAGTGTCCAGATCAGTGACACTCTGTCTGGCTTACTTGGTCAAATGGCAGAAGATGACCCTACGTGATGCCTACCACTACTTGAAGATAAGACGACCACAAATTCGACCGAACACGGGTTTCTTCAAGCAGCTCATCAAATTCGAAGAAAGACTGTTCAGCGAAGCAACAGTCAAGATGGTGTATTGCGAGGCCATCGACAAAGAGATCCCTGATGTATACGAGCCTGATTACAGCGGAATGACATGGTTCAGGCAACGATACGGACCTATCGAAAAATGCTGA
Protein
MKVLTVGDKVASVGRENEPTVVVDTQKLIREELNRGCPLGVSRVTNLVYVCGAHALPGAVRVLHPGLIVSAAPELPPPPEDYVPRHFVPLLDTPNSDMLPYMDRVADLINEVVTNGGVVLVHCVAGVSRSVTLCLAYLVKWQKMTLRDAYHYLKIRRPQIRPNTGFFKQLIKFEERLFSEATVKMVYCEAIDKEIPDVYEPDYSGMTWFRQRYGPIEKC
Summary
Similarity
Belongs to the protein-tyrosine phosphatase family. Non-receptor class dual specificity subfamily.
Uniprot
H9JMV2
A0A0L7KUI1
A0A2H1X2U7
A0A2A4J7E1
A0A0N0PBN7
A0A212F019
+ More
A0A194Q3P1 W8B462 A0A084WGG7 Q7Q581 A0A182Y2L3 A0A182LKE9 A0A182I4H0 A0A182MPX2 A0A182J2Z9 A0A0K8V8S5 A0A182X4H3 A0A182UWF9 A0A0K8VBP4 A0A182NIB2 A0A182K5K6 A0A182PMT1 A0A182R942 A0A182R8U7 A0A1I8N644 A0A1I8MCA1 A0A182QHS0 A0A182TIN9 B4JES1 A0A1I8PBT7 A0A034VST2 A0A3B0KBJ6 A0A182FEW9 B4M6I2 T1PCJ3 A0A2M4DS72 A0A0A1WZ98 A0A182WBS1 A0A0L0BU36 D0QWR1 B4NJK0 A0A1L8DKQ6 W5J648 A0A226DZZ4 A0A1L8E3D4 B4GNI3 Q29C71 B4K804 A0A1J1J2M0 Q9VAB0 T1DKC2 B4R1H7 B4HZL3 B3P7Z2 B4PLR9 Q17DB5 A0A1Y1KH41 A0A1B0CIK7 A0A0M5J6X0 A0A1W4VHS5 T1IQY8 A0A0K8SUG0 T1JMK9 A0A1B0GPA2 D3TQ52 A0A1A9VHC4 A0A1A9Z959 B7P362 A0A1W2WBL1 F6W4Z8 A0A131Y026 V5IGC6 A0A1B0BRR4 A0A1S3IP06 A0A210QAZ4
A0A194Q3P1 W8B462 A0A084WGG7 Q7Q581 A0A182Y2L3 A0A182LKE9 A0A182I4H0 A0A182MPX2 A0A182J2Z9 A0A0K8V8S5 A0A182X4H3 A0A182UWF9 A0A0K8VBP4 A0A182NIB2 A0A182K5K6 A0A182PMT1 A0A182R942 A0A182R8U7 A0A1I8N644 A0A1I8MCA1 A0A182QHS0 A0A182TIN9 B4JES1 A0A1I8PBT7 A0A034VST2 A0A3B0KBJ6 A0A182FEW9 B4M6I2 T1PCJ3 A0A2M4DS72 A0A0A1WZ98 A0A182WBS1 A0A0L0BU36 D0QWR1 B4NJK0 A0A1L8DKQ6 W5J648 A0A226DZZ4 A0A1L8E3D4 B4GNI3 Q29C71 B4K804 A0A1J1J2M0 Q9VAB0 T1DKC2 B4R1H7 B4HZL3 B3P7Z2 B4PLR9 Q17DB5 A0A1Y1KH41 A0A1B0CIK7 A0A0M5J6X0 A0A1W4VHS5 T1IQY8 A0A0K8SUG0 T1JMK9 A0A1B0GPA2 D3TQ52 A0A1A9VHC4 A0A1A9Z959 B7P362 A0A1W2WBL1 F6W4Z8 A0A131Y026 V5IGC6 A0A1B0BRR4 A0A1S3IP06 A0A210QAZ4
Pubmed
19121390
26227816
26354079
22118469
24495485
24438588
+ More
12364791 14747013 17210077 25244985 20966253 25315136 17994087 25348373 25830018 26108605 19859648 20920257 23761445 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 17510324 28004739 26823975 20353571 12481130 15114417 25765539 28812685
12364791 14747013 17210077 25244985 20966253 25315136 17994087 25348373 25830018 26108605 19859648 20920257 23761445 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 17510324 28004739 26823975 20353571 12481130 15114417 25765539 28812685
EMBL
BABH01029151
JTDY01005488
KOB66952.1
ODYU01012980
SOQ59558.1
NWSH01002601
+ More
PCG67881.1 KQ460874 KPJ11665.1 AGBW02011206 OWR47057.1 KQ459586 KPI98030.1 GAMC01013193 JAB93362.1 ATLV01023568 KE525344 KFB49311.1 AAAB01008960 EAA11667.3 APCN01002312 AXCM01000123 GDHF01017017 JAI35297.1 GDHF01016048 JAI36266.1 AXCN02000879 CH916369 EDV93202.1 GAKP01013433 GAKP01013432 JAC45519.1 OUUW01000005 SPP80948.1 CH940652 EDW59258.1 KA646472 AFP61101.1 GGFL01016191 MBW80369.1 GBXI01009913 GBXI01001297 JAD04379.1 JAD12995.1 JRES01001347 KNC23498.1 FJ821034 ACN94752.1 CH964272 EDW83924.1 GFDF01007035 JAV07049.1 ADMH02002146 ETN58284.1 LNIX01000009 OXA50374.1 GFDF01000992 JAV13092.1 CH479186 EDW39309.1 CM000070 EAL26775.2 CH933806 EDW14338.1 CVRI01000067 CRL06608.1 AE014297 BT032968 AAF57003.2 ACD99532.1 GAMD01001011 JAB00580.1 CM000364 EDX14965.1 CH480819 EDW53470.1 CH954182 EDV53119.1 CM000160 EDW99056.2 CH477297 EAT44375.1 GEZM01088611 JAV58067.1 AJWK01013496 CP012526 ALC46256.1 AFFK01018426 GBRD01008967 GDHC01019428 JAG56854.1 JAP99200.1 JH432105 AJVK01033253 CCAG010022183 EZ423554 ADD19830.1 ABJB010256539 DS626813 EEC01034.1 EAAA01000286 GEFM01003177 JAP72619.1 GANP01006922 JAB77546.1 JXJN01019253 JXJN01019254 NEDP02004373 OWF45899.1
PCG67881.1 KQ460874 KPJ11665.1 AGBW02011206 OWR47057.1 KQ459586 KPI98030.1 GAMC01013193 JAB93362.1 ATLV01023568 KE525344 KFB49311.1 AAAB01008960 EAA11667.3 APCN01002312 AXCM01000123 GDHF01017017 JAI35297.1 GDHF01016048 JAI36266.1 AXCN02000879 CH916369 EDV93202.1 GAKP01013433 GAKP01013432 JAC45519.1 OUUW01000005 SPP80948.1 CH940652 EDW59258.1 KA646472 AFP61101.1 GGFL01016191 MBW80369.1 GBXI01009913 GBXI01001297 JAD04379.1 JAD12995.1 JRES01001347 KNC23498.1 FJ821034 ACN94752.1 CH964272 EDW83924.1 GFDF01007035 JAV07049.1 ADMH02002146 ETN58284.1 LNIX01000009 OXA50374.1 GFDF01000992 JAV13092.1 CH479186 EDW39309.1 CM000070 EAL26775.2 CH933806 EDW14338.1 CVRI01000067 CRL06608.1 AE014297 BT032968 AAF57003.2 ACD99532.1 GAMD01001011 JAB00580.1 CM000364 EDX14965.1 CH480819 EDW53470.1 CH954182 EDV53119.1 CM000160 EDW99056.2 CH477297 EAT44375.1 GEZM01088611 JAV58067.1 AJWK01013496 CP012526 ALC46256.1 AFFK01018426 GBRD01008967 GDHC01019428 JAG56854.1 JAP99200.1 JH432105 AJVK01033253 CCAG010022183 EZ423554 ADD19830.1 ABJB010256539 DS626813 EEC01034.1 EAAA01000286 GEFM01003177 JAP72619.1 GANP01006922 JAB77546.1 JXJN01019253 JXJN01019254 NEDP02004373 OWF45899.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000007151
UP000053268
+ More
UP000030765 UP000007062 UP000076408 UP000075882 UP000075840 UP000075883 UP000075880 UP000076407 UP000075903 UP000075884 UP000075881 UP000075885 UP000075900 UP000095301 UP000075886 UP000075902 UP000001070 UP000095300 UP000268350 UP000069272 UP000008792 UP000075920 UP000037069 UP000007798 UP000000673 UP000198287 UP000008744 UP000001819 UP000009192 UP000183832 UP000000803 UP000000304 UP000001292 UP000008711 UP000002282 UP000008820 UP000092461 UP000092553 UP000192221 UP000092462 UP000092444 UP000078200 UP000092445 UP000001555 UP000008144 UP000092460 UP000085678 UP000242188
UP000030765 UP000007062 UP000076408 UP000075882 UP000075840 UP000075883 UP000075880 UP000076407 UP000075903 UP000075884 UP000075881 UP000075885 UP000075900 UP000095301 UP000075886 UP000075902 UP000001070 UP000095300 UP000268350 UP000069272 UP000008792 UP000075920 UP000037069 UP000007798 UP000000673 UP000198287 UP000008744 UP000001819 UP000009192 UP000183832 UP000000803 UP000000304 UP000001292 UP000008711 UP000002282 UP000008820 UP000092461 UP000092553 UP000192221 UP000092462 UP000092444 UP000078200 UP000092445 UP000001555 UP000008144 UP000092460 UP000085678 UP000242188
Pfam
PF00782 DSPc
Interpro
SUPFAM
SSF52799
SSF52799
Gene 3D
CDD
ProteinModelPortal
H9JMV2
A0A0L7KUI1
A0A2H1X2U7
A0A2A4J7E1
A0A0N0PBN7
A0A212F019
+ More
A0A194Q3P1 W8B462 A0A084WGG7 Q7Q581 A0A182Y2L3 A0A182LKE9 A0A182I4H0 A0A182MPX2 A0A182J2Z9 A0A0K8V8S5 A0A182X4H3 A0A182UWF9 A0A0K8VBP4 A0A182NIB2 A0A182K5K6 A0A182PMT1 A0A182R942 A0A182R8U7 A0A1I8N644 A0A1I8MCA1 A0A182QHS0 A0A182TIN9 B4JES1 A0A1I8PBT7 A0A034VST2 A0A3B0KBJ6 A0A182FEW9 B4M6I2 T1PCJ3 A0A2M4DS72 A0A0A1WZ98 A0A182WBS1 A0A0L0BU36 D0QWR1 B4NJK0 A0A1L8DKQ6 W5J648 A0A226DZZ4 A0A1L8E3D4 B4GNI3 Q29C71 B4K804 A0A1J1J2M0 Q9VAB0 T1DKC2 B4R1H7 B4HZL3 B3P7Z2 B4PLR9 Q17DB5 A0A1Y1KH41 A0A1B0CIK7 A0A0M5J6X0 A0A1W4VHS5 T1IQY8 A0A0K8SUG0 T1JMK9 A0A1B0GPA2 D3TQ52 A0A1A9VHC4 A0A1A9Z959 B7P362 A0A1W2WBL1 F6W4Z8 A0A131Y026 V5IGC6 A0A1B0BRR4 A0A1S3IP06 A0A210QAZ4
A0A194Q3P1 W8B462 A0A084WGG7 Q7Q581 A0A182Y2L3 A0A182LKE9 A0A182I4H0 A0A182MPX2 A0A182J2Z9 A0A0K8V8S5 A0A182X4H3 A0A182UWF9 A0A0K8VBP4 A0A182NIB2 A0A182K5K6 A0A182PMT1 A0A182R942 A0A182R8U7 A0A1I8N644 A0A1I8MCA1 A0A182QHS0 A0A182TIN9 B4JES1 A0A1I8PBT7 A0A034VST2 A0A3B0KBJ6 A0A182FEW9 B4M6I2 T1PCJ3 A0A2M4DS72 A0A0A1WZ98 A0A182WBS1 A0A0L0BU36 D0QWR1 B4NJK0 A0A1L8DKQ6 W5J648 A0A226DZZ4 A0A1L8E3D4 B4GNI3 Q29C71 B4K804 A0A1J1J2M0 Q9VAB0 T1DKC2 B4R1H7 B4HZL3 B3P7Z2 B4PLR9 Q17DB5 A0A1Y1KH41 A0A1B0CIK7 A0A0M5J6X0 A0A1W4VHS5 T1IQY8 A0A0K8SUG0 T1JMK9 A0A1B0GPA2 D3TQ52 A0A1A9VHC4 A0A1A9Z959 B7P362 A0A1W2WBL1 F6W4Z8 A0A131Y026 V5IGC6 A0A1B0BRR4 A0A1S3IP06 A0A210QAZ4
PDB
2WGP
E-value=3.97817e-27,
Score=299
Ontologies
GO
Topology
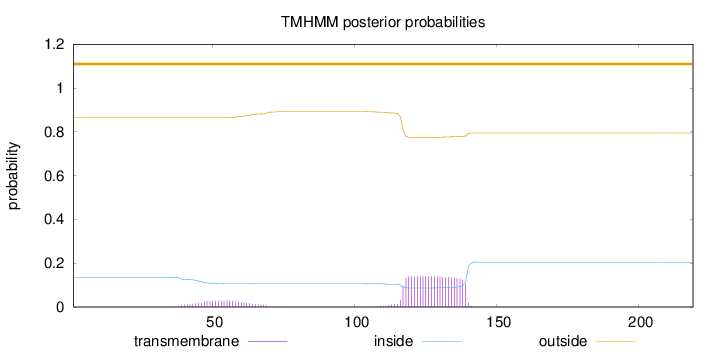
Length:
219
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.79423
Exp number, first 60 AAs:
0.48252
Total prob of N-in:
0.13574
outside
1 - 219
Population Genetic Test Statistics
Pi
21.083867
Theta
21.374597
Tajima's D
0.003626
CLR
0.194126
CSRT
0.373881305934703
Interpretation
Uncertain
