Gene
KWMTBOMO13407
Pre Gene Modal
BGIBMGA010851
Annotation
Uncharacterized_protein_OBRU01_07690_[Operophtera_brumata]
Location in the cell
PlasmaMembrane Reliability : 4.567
Sequence
CDS
ATGGCTGCGCTTAGGGTGTTTAGCCGTAATCTTCCGTATTTGCGACAACAATTTGCAGCATTGTTAGGGAACACATCACCTTCGGTAAAGTTTATCTGCGTGGTCGTGGTTATAGGATATATATTATCATTTTCCGATGATGTCGTCAATGTACTCAGTGTCACTCCAGGGTATTTGCTGCCTCCTTCATTTCAACTATGGTCTACTTTCACGTTCTGGTTTCTCGAAATACATTTGTGGGAGGTGATCATCGATATTGTGACTGTGGGGCTGTGCGGGAAACTCATAGAACCACTGTGGGGACAGATGGAAATGATGAAGTTTTTCATACTGACCAACTTTGGTGTAGCGTTTTTGACTACGTTTTACTATTTAGTGATATTTGCGTGTACGGGTCAAACCTCGTATTTATTCGACATTCATGTACACGGGCTGGCAGGGTACCTGGCCGCAGTCAGTGTCGCTGTGAAGCAAATCATGCCAGATCATCTGCTGATTAAAACACCACTAGGTAAATTAACCAACAGGTCTTTACCACTACTGATCCTAATAGTGGCTATTATCCTGTGGGCTATTGGTGCCTTAGAAGGGACATATCCTTGCATGTGGGGCAGTGGGACGATTCTATCTTGGATATATCTAAGGTTCTGGCAAAGACACACCAATGGTACCAGGGGAGATATGGCAGACAACTTTAGTTTTGATAACTTCTTTCCAACAGTCCTGCAACCTGTAGTCCGAGCTATTGTAGGTCCACCGCATCGTTGTCTAGTGCGGCTTGGTCTCTGTTCACCTAGTCCACGTCGAGTGCACTTAGCACTTAGTCCCCGAGGTGTGACAATTTCTATGCCTGGAGTGGAACCACAAGATATGGAGAGGCGAAGACAAATTGCATTAAAAGCATTGAGTGAAAGACTATCCAAAACATCTGAGCAAACTCGTCAAAAGAACTTATCGACGAAAGTAAACATGGATGCTGTTAGGAAGATGCAGACATCACAAGTGATACCTCAATTCACGGAACCCGAAACAAGCAAAGGTATACAGGCGCCTCCCACGGAGGCGACCCTTGTGGATCTCGGACCAGACCCAACACCTCCAATCACCAAGCAATCATGA
Protein
MAALRVFSRNLPYLRQQFAALLGNTSPSVKFICVVVVIGYILSFSDDVVNVLSVTPGYLLPPSFQLWSTFTFWFLEIHLWEVIIDIVTVGLCGKLIEPLWGQMEMMKFFILTNFGVAFLTTFYYLVIFACTGQTSYLFDIHVHGLAGYLAAVSVAVKQIMPDHLLIKTPLGKLTNRSLPLLILIVAIILWAIGALEGTYPCMWGSGTILSWIYLRFWQRHTNGTRGDMADNFSFDNFFPTVLQPVVRAIVGPPHRCLVRLGLCSPSPRRVHLALSPRGVTISMPGVEPQDMERRRQIALKALSERLSKTSEQTRQKNLSTKVNMDAVRKMQTSQVIPQFTEPETSKGIQAPPTEATLVDLGPDPTPPITKQS
Summary
Uniprot
H9JMU8
A0A2H1VBE3
A0A0L7LJN5
A0A194RDM1
A0A194Q3N6
A0A212ES39
+ More
A0A2J7RHU6 A0A2P8XJ93 A0A1W4XUI8 D6WZF9 A0A1B6KNC7 A0A0M8ZSD5 A0A0L7QWX2 A0A067RRC2 E2BNW4 K7ISP7 A0A1W4XK18 A0A088AD27 A0A1B6H8N4 A0A2A3EM74 A0A026WZC0 A0A1B6ETB8 E2AFT0 A0A151X5L9 A0A158NSV2 A0A195EXB8 A0A151I0K2 A0A195DPR0 F4WSJ6 A0A1B6D489 E9IPX5 A0A195CT02 A0A0J7L0K8 A0A0A9WFT4 A0A1Y1L4V0 N6UEU0 A0A0C9RWX6 A0A0P4W4P6 T1HHS6 Q0IFM8 A0A1S4F7R8 A0A0N8D8K4 A0A023EU16 A0A182GWA9 A0A0P5G1G5 A0A0N8CFY1 E0VAX0 E9GJT9 A0A0P5V1V8 A0A1Q3F6K1 A0A224XRE2 A0A0P5V2H8 A0A0K8TMY3 A0A1I8PM05 T1E1H4 W5J912 A0A2M4AEV8 A0A2M4AEA8 A0A2M4AEC5 A0A2M4BMH8 A0A2M4BMI1 A0A2M4BMK4 D3TQJ0 A0A182THV3 A0A182V8L5 A0A2M3ZGV5 A0A182VUT7 A0A182XIP2 A0A1S4H134 Q7QKG4 A0A182JH08 A0A182HG45 A0A1A9W5H8 A0A2M3Z3Z2 A0A182F5A5 A0A2M3Z410 A0A0P6I9E7 A0A0M3QU83 A0A182QAV9 B4KKX6 A0A0N8DL14 A0A182NEK7 A0A1I8MPI8 T1PJ24 B4M9K2 B0W242 B0X6D6 A0A084VPS1 A0A182M801 A0A182RGA2 A0A182K1L6 A0A0P5G6K1 A0A182YFZ1 Q29PD9 A0A0K8UWN0 B4GKK4 A0A0P5Y2B8 A0A3B0KBH3 A0A034VLR7 A0A1B0BN35
A0A2J7RHU6 A0A2P8XJ93 A0A1W4XUI8 D6WZF9 A0A1B6KNC7 A0A0M8ZSD5 A0A0L7QWX2 A0A067RRC2 E2BNW4 K7ISP7 A0A1W4XK18 A0A088AD27 A0A1B6H8N4 A0A2A3EM74 A0A026WZC0 A0A1B6ETB8 E2AFT0 A0A151X5L9 A0A158NSV2 A0A195EXB8 A0A151I0K2 A0A195DPR0 F4WSJ6 A0A1B6D489 E9IPX5 A0A195CT02 A0A0J7L0K8 A0A0A9WFT4 A0A1Y1L4V0 N6UEU0 A0A0C9RWX6 A0A0P4W4P6 T1HHS6 Q0IFM8 A0A1S4F7R8 A0A0N8D8K4 A0A023EU16 A0A182GWA9 A0A0P5G1G5 A0A0N8CFY1 E0VAX0 E9GJT9 A0A0P5V1V8 A0A1Q3F6K1 A0A224XRE2 A0A0P5V2H8 A0A0K8TMY3 A0A1I8PM05 T1E1H4 W5J912 A0A2M4AEV8 A0A2M4AEA8 A0A2M4AEC5 A0A2M4BMH8 A0A2M4BMI1 A0A2M4BMK4 D3TQJ0 A0A182THV3 A0A182V8L5 A0A2M3ZGV5 A0A182VUT7 A0A182XIP2 A0A1S4H134 Q7QKG4 A0A182JH08 A0A182HG45 A0A1A9W5H8 A0A2M3Z3Z2 A0A182F5A5 A0A2M3Z410 A0A0P6I9E7 A0A0M3QU83 A0A182QAV9 B4KKX6 A0A0N8DL14 A0A182NEK7 A0A1I8MPI8 T1PJ24 B4M9K2 B0W242 B0X6D6 A0A084VPS1 A0A182M801 A0A182RGA2 A0A182K1L6 A0A0P5G6K1 A0A182YFZ1 Q29PD9 A0A0K8UWN0 B4GKK4 A0A0P5Y2B8 A0A3B0KBH3 A0A034VLR7 A0A1B0BN35
Pubmed
19121390
26227816
26354079
22118469
29403074
18362917
+ More
19820115 24845553 20798317 20075255 24508170 30249741 21347285 21719571 21282665 25401762 26823975 28004739 23537049 27129103 17510324 24945155 26483478 20566863 21292972 26369729 24330624 20920257 23761445 20353571 12364791 17994087 25315136 24438588 25244985 15632085 25348373
19820115 24845553 20798317 20075255 24508170 30249741 21347285 21719571 21282665 25401762 26823975 28004739 23537049 27129103 17510324 24945155 26483478 20566863 21292972 26369729 24330624 20920257 23761445 20353571 12364791 17994087 25315136 24438588 25244985 15632085 25348373
EMBL
BABH01029173
ODYU01001647
SOQ38163.1
JTDY01000878
KOB75579.1
KQ460323
+ More
KPJ15938.1 KQ459586 KPI98025.1 AGBW02012888 OWR44279.1 NEVH01003737 PNF40396.1 PYGN01001944 PSN32071.1 KQ971372 EFA10421.1 GEBQ01027046 JAT12931.1 KQ435878 KOX69987.1 KQ414706 KOC63066.1 KK852511 KDR22304.1 GL449511 EFN82556.1 AAZX01002898 GECU01036686 JAS71020.1 KZ288220 PBC32141.1 KK107054 QOIP01000004 EZA61415.1 RLU24008.1 GECZ01028806 JAS40963.1 GL439158 EFN67679.1 KQ982494 KYQ55701.1 ADTU01025191 KQ981923 KYN32925.1 KQ976619 KYM79129.1 KQ980653 KYN14836.1 GL888322 EGI62811.1 GEDC01016832 JAS20466.1 GL764659 EFZ17348.1 KQ977305 KYN03775.1 LBMM01001604 KMQ96029.1 GBHO01043045 GBHO01039887 GBHO01039884 GBHO01039880 GBHO01037834 GBHO01037830 GBHO01037829 GBHO01035866 GBHO01019446 GBHO01009894 GBHO01009291 GDHC01020381 JAG00559.1 JAG03717.1 JAG03720.1 JAG03724.1 JAG05770.1 JAG05774.1 JAG05775.1 JAG07738.1 JAG24158.1 JAG33710.1 JAG34313.1 JAP98247.1 GEZM01064570 JAV68712.1 APGK01029446 KB740735 KB630388 KB632136 ENN79126.1 ERL83585.1 ERL89063.1 GBYB01012366 JAG82133.1 GDKW01000280 JAI56315.1 ACPB03001010 CH477306 EAT44115.1 GDIP01058215 JAM45500.1 GAPW01001664 JAC11934.1 JXUM01092900 KQ564018 KXJ72928.1 GDIQ01247653 GDIP01067966 LRGB01001363 JAK04072.1 JAM35749.1 KZS12180.1 GDIP01135599 JAL68115.1 DS235019 EEB10526.1 GL732548 EFX80170.1 GDIP01105650 JAL98064.1 GFDL01011855 JAV23190.1 GFTR01005416 JAW11010.1 GDIP01105651 JAL98063.1 GDAI01002107 JAI15496.1 GALA01001664 JAA93188.1 ADMH02002093 ETN59289.1 GGFK01005847 MBW39168.1 GGFK01005804 MBW39125.1 GGFK01005816 MBW39137.1 GGFJ01005145 MBW54286.1 GGFJ01005146 MBW54287.1 GGFJ01005144 MBW54285.1 EZ423692 ADD19968.1 GGFM01007001 MBW27752.1 AAAB01008797 EAA03623.3 AXCP01008030 APCN01004115 GGFM01002417 MBW23168.1 GGFM01002437 MBW23188.1 GDIQ01007699 JAN87038.1 CP012523 ALC40213.1 AXCN02000851 CH933807 EDW11706.1 GDIP01023335 JAM80380.1 KA647913 AFP62542.1 CH940654 EDW57878.1 DS231825 EDS28170.1 DS232411 EDS41286.1 ATLV01015034 KE524999 KFB39965.1 AXCM01000484 GDIQ01246554 JAK05171.1 CH379058 EAL34354.2 GDHF01021318 JAI30996.1 CH479184 EDW37170.1 GDIP01077147 JAM26568.1 OUUW01000064 SPP90062.1 GAKP01016232 JAC42720.1 JXJN01017195
KPJ15938.1 KQ459586 KPI98025.1 AGBW02012888 OWR44279.1 NEVH01003737 PNF40396.1 PYGN01001944 PSN32071.1 KQ971372 EFA10421.1 GEBQ01027046 JAT12931.1 KQ435878 KOX69987.1 KQ414706 KOC63066.1 KK852511 KDR22304.1 GL449511 EFN82556.1 AAZX01002898 GECU01036686 JAS71020.1 KZ288220 PBC32141.1 KK107054 QOIP01000004 EZA61415.1 RLU24008.1 GECZ01028806 JAS40963.1 GL439158 EFN67679.1 KQ982494 KYQ55701.1 ADTU01025191 KQ981923 KYN32925.1 KQ976619 KYM79129.1 KQ980653 KYN14836.1 GL888322 EGI62811.1 GEDC01016832 JAS20466.1 GL764659 EFZ17348.1 KQ977305 KYN03775.1 LBMM01001604 KMQ96029.1 GBHO01043045 GBHO01039887 GBHO01039884 GBHO01039880 GBHO01037834 GBHO01037830 GBHO01037829 GBHO01035866 GBHO01019446 GBHO01009894 GBHO01009291 GDHC01020381 JAG00559.1 JAG03717.1 JAG03720.1 JAG03724.1 JAG05770.1 JAG05774.1 JAG05775.1 JAG07738.1 JAG24158.1 JAG33710.1 JAG34313.1 JAP98247.1 GEZM01064570 JAV68712.1 APGK01029446 KB740735 KB630388 KB632136 ENN79126.1 ERL83585.1 ERL89063.1 GBYB01012366 JAG82133.1 GDKW01000280 JAI56315.1 ACPB03001010 CH477306 EAT44115.1 GDIP01058215 JAM45500.1 GAPW01001664 JAC11934.1 JXUM01092900 KQ564018 KXJ72928.1 GDIQ01247653 GDIP01067966 LRGB01001363 JAK04072.1 JAM35749.1 KZS12180.1 GDIP01135599 JAL68115.1 DS235019 EEB10526.1 GL732548 EFX80170.1 GDIP01105650 JAL98064.1 GFDL01011855 JAV23190.1 GFTR01005416 JAW11010.1 GDIP01105651 JAL98063.1 GDAI01002107 JAI15496.1 GALA01001664 JAA93188.1 ADMH02002093 ETN59289.1 GGFK01005847 MBW39168.1 GGFK01005804 MBW39125.1 GGFK01005816 MBW39137.1 GGFJ01005145 MBW54286.1 GGFJ01005146 MBW54287.1 GGFJ01005144 MBW54285.1 EZ423692 ADD19968.1 GGFM01007001 MBW27752.1 AAAB01008797 EAA03623.3 AXCP01008030 APCN01004115 GGFM01002417 MBW23168.1 GGFM01002437 MBW23188.1 GDIQ01007699 JAN87038.1 CP012523 ALC40213.1 AXCN02000851 CH933807 EDW11706.1 GDIP01023335 JAM80380.1 KA647913 AFP62542.1 CH940654 EDW57878.1 DS231825 EDS28170.1 DS232411 EDS41286.1 ATLV01015034 KE524999 KFB39965.1 AXCM01000484 GDIQ01246554 JAK05171.1 CH379058 EAL34354.2 GDHF01021318 JAI30996.1 CH479184 EDW37170.1 GDIP01077147 JAM26568.1 OUUW01000064 SPP90062.1 GAKP01016232 JAC42720.1 JXJN01017195
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000007151
UP000235965
+ More
UP000245037 UP000192223 UP000007266 UP000053105 UP000053825 UP000027135 UP000008237 UP000002358 UP000005203 UP000242457 UP000053097 UP000279307 UP000000311 UP000075809 UP000005205 UP000078541 UP000078540 UP000078492 UP000007755 UP000078542 UP000036403 UP000019118 UP000030742 UP000015103 UP000008820 UP000069940 UP000249989 UP000076858 UP000009046 UP000000305 UP000095300 UP000000673 UP000075902 UP000075903 UP000075920 UP000076407 UP000007062 UP000075880 UP000075840 UP000091820 UP000069272 UP000092553 UP000075886 UP000009192 UP000075884 UP000095301 UP000008792 UP000002320 UP000030765 UP000075883 UP000075900 UP000075881 UP000076408 UP000001819 UP000008744 UP000268350 UP000092460
UP000245037 UP000192223 UP000007266 UP000053105 UP000053825 UP000027135 UP000008237 UP000002358 UP000005203 UP000242457 UP000053097 UP000279307 UP000000311 UP000075809 UP000005205 UP000078541 UP000078540 UP000078492 UP000007755 UP000078542 UP000036403 UP000019118 UP000030742 UP000015103 UP000008820 UP000069940 UP000249989 UP000076858 UP000009046 UP000000305 UP000095300 UP000000673 UP000075902 UP000075903 UP000075920 UP000076407 UP000007062 UP000075880 UP000075840 UP000091820 UP000069272 UP000092553 UP000075886 UP000009192 UP000075884 UP000095301 UP000008792 UP000002320 UP000030765 UP000075883 UP000075900 UP000075881 UP000076408 UP000001819 UP000008744 UP000268350 UP000092460
PRIDE
Interpro
SUPFAM
SSF54160
SSF54160
Gene 3D
CDD
ProteinModelPortal
H9JMU8
A0A2H1VBE3
A0A0L7LJN5
A0A194RDM1
A0A194Q3N6
A0A212ES39
+ More
A0A2J7RHU6 A0A2P8XJ93 A0A1W4XUI8 D6WZF9 A0A1B6KNC7 A0A0M8ZSD5 A0A0L7QWX2 A0A067RRC2 E2BNW4 K7ISP7 A0A1W4XK18 A0A088AD27 A0A1B6H8N4 A0A2A3EM74 A0A026WZC0 A0A1B6ETB8 E2AFT0 A0A151X5L9 A0A158NSV2 A0A195EXB8 A0A151I0K2 A0A195DPR0 F4WSJ6 A0A1B6D489 E9IPX5 A0A195CT02 A0A0J7L0K8 A0A0A9WFT4 A0A1Y1L4V0 N6UEU0 A0A0C9RWX6 A0A0P4W4P6 T1HHS6 Q0IFM8 A0A1S4F7R8 A0A0N8D8K4 A0A023EU16 A0A182GWA9 A0A0P5G1G5 A0A0N8CFY1 E0VAX0 E9GJT9 A0A0P5V1V8 A0A1Q3F6K1 A0A224XRE2 A0A0P5V2H8 A0A0K8TMY3 A0A1I8PM05 T1E1H4 W5J912 A0A2M4AEV8 A0A2M4AEA8 A0A2M4AEC5 A0A2M4BMH8 A0A2M4BMI1 A0A2M4BMK4 D3TQJ0 A0A182THV3 A0A182V8L5 A0A2M3ZGV5 A0A182VUT7 A0A182XIP2 A0A1S4H134 Q7QKG4 A0A182JH08 A0A182HG45 A0A1A9W5H8 A0A2M3Z3Z2 A0A182F5A5 A0A2M3Z410 A0A0P6I9E7 A0A0M3QU83 A0A182QAV9 B4KKX6 A0A0N8DL14 A0A182NEK7 A0A1I8MPI8 T1PJ24 B4M9K2 B0W242 B0X6D6 A0A084VPS1 A0A182M801 A0A182RGA2 A0A182K1L6 A0A0P5G6K1 A0A182YFZ1 Q29PD9 A0A0K8UWN0 B4GKK4 A0A0P5Y2B8 A0A3B0KBH3 A0A034VLR7 A0A1B0BN35
A0A2J7RHU6 A0A2P8XJ93 A0A1W4XUI8 D6WZF9 A0A1B6KNC7 A0A0M8ZSD5 A0A0L7QWX2 A0A067RRC2 E2BNW4 K7ISP7 A0A1W4XK18 A0A088AD27 A0A1B6H8N4 A0A2A3EM74 A0A026WZC0 A0A1B6ETB8 E2AFT0 A0A151X5L9 A0A158NSV2 A0A195EXB8 A0A151I0K2 A0A195DPR0 F4WSJ6 A0A1B6D489 E9IPX5 A0A195CT02 A0A0J7L0K8 A0A0A9WFT4 A0A1Y1L4V0 N6UEU0 A0A0C9RWX6 A0A0P4W4P6 T1HHS6 Q0IFM8 A0A1S4F7R8 A0A0N8D8K4 A0A023EU16 A0A182GWA9 A0A0P5G1G5 A0A0N8CFY1 E0VAX0 E9GJT9 A0A0P5V1V8 A0A1Q3F6K1 A0A224XRE2 A0A0P5V2H8 A0A0K8TMY3 A0A1I8PM05 T1E1H4 W5J912 A0A2M4AEV8 A0A2M4AEA8 A0A2M4AEC5 A0A2M4BMH8 A0A2M4BMI1 A0A2M4BMK4 D3TQJ0 A0A182THV3 A0A182V8L5 A0A2M3ZGV5 A0A182VUT7 A0A182XIP2 A0A1S4H134 Q7QKG4 A0A182JH08 A0A182HG45 A0A1A9W5H8 A0A2M3Z3Z2 A0A182F5A5 A0A2M3Z410 A0A0P6I9E7 A0A0M3QU83 A0A182QAV9 B4KKX6 A0A0N8DL14 A0A182NEK7 A0A1I8MPI8 T1PJ24 B4M9K2 B0W242 B0X6D6 A0A084VPS1 A0A182M801 A0A182RGA2 A0A182K1L6 A0A0P5G6K1 A0A182YFZ1 Q29PD9 A0A0K8UWN0 B4GKK4 A0A0P5Y2B8 A0A3B0KBH3 A0A034VLR7 A0A1B0BN35
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
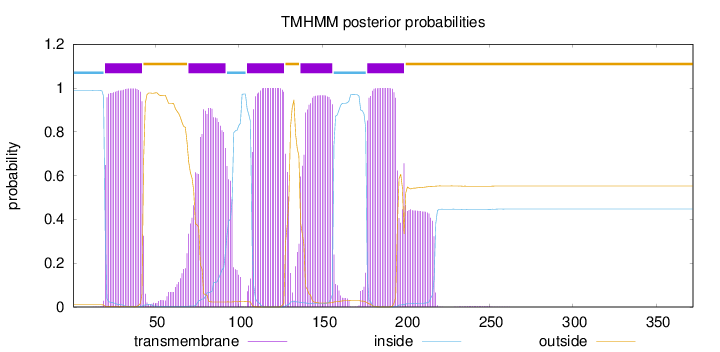
Length:
372
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
114.43676
Exp number, first 60 AAs:
23.26175
Total prob of N-in:
0.98968
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 42
outside
43 - 69
TMhelix
70 - 92
inside
93 - 104
TMhelix
105 - 127
outside
128 - 136
TMhelix
137 - 156
inside
157 - 176
TMhelix
177 - 199
outside
200 - 372
Population Genetic Test Statistics
Pi
15.935084
Theta
17.754916
Tajima's D
-0.328176
CLR
0.688482
CSRT
0.290085495725214
Interpretation
Uncertain
