Gene
KWMTBOMO13405 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010906
Annotation
PREDICTED:_proliferating_cell_nuclear_antigen_isoform_X1_[Bombyx_mori]
Full name
Proliferating cell nuclear antigen
Alternative Name
Cyclin
Mutagen-sensitive 209 protein
Mutagen-sensitive 209 protein
Location in the cell
Nuclear Reliability : 4.469
Sequence
CDS
ATGTTTGAGGCACGTTTACTCCGAAGCTCTATCTTAAAGAAGGTCTTGGAGGCTATTAAAGATCTGCTGACACAGGCCACTTTCGATTGCGATGACAATGGAATTCAGTTACAGGCTATGGACAACTCTCATGTGTCTCTCGTGTCACTCACTCTCCGAGCTGATGGCTTTGACAAGTACCGCTGCGATAGGAACATCTCAATGGGCATGAATCTAGGCAGCATGTCAAAGATTCTCAAATGTGCTGGAGATAAGGATACAGTTACAATAAAAGCACAGGATAATGCTGACAATGTCACATTTGTTTTTGAGAGCCCAAATCAAGAGAAAGTCTCTGATTACGAGATGAAGCTTATGAATTTGGATCTTGAACATTTAGGTATTCCAGAGACTGAATACAGCTGCACTATTCGCATGCCAAGTTCTGAATTTGCTAGAATCTGCCGGGATCTCTCACAGTTTGGAGAATCAATGGTGATTTCATGCACAAAAGAAGGAGTAAAGTTCTCGGCAACAGGCGACATCGGCTCAGCGAACGTCAAGCTGGCCCAGACCGCTTCTATTGACAAAGAGGAAGAGGCAGTCGTCATTGAAATGGAAGAGCCCGTCACTCTGACGTTTGCCTGCCAGTACCTCAACTATTTCACTAAAGCTACTTCTCTCAGCCCTCAGGTGCAGCTGTCGATGTCAGCAGACGTTCCTTTGGTGGTGGAGTACCGCATCCCGGACATTGGCCACATCCGCTACTACCTGGCGCCTAAGATCGAGGAAGAAGACAGCTGA
Protein
MFEARLLRSSILKKVLEAIKDLLTQATFDCDDNGIQLQAMDNSHVSLVSLTLRADGFDKYRCDRNISMGMNLGSMSKILKCAGDKDTVTIKAQDNADNVTFVFESPNQEKVSDYEMKLMNLDLEHLGIPETEYSCTIRMPSSEFARICRDLSQFGESMVISCTKEGVKFSATGDIGSANVKLAQTASIDKEEEAVVIEMEEPVTLTFACQYLNYFTKATSLSPQVQLSMSADVPLVVEYRIPDIGHIRYYLAPKIEEEDS
Summary
Description
This protein is an auxiliary protein of DNA polymerase delta and is involved in the control of eukaryotic DNA replication by increasing the polymerase's processibility during elongation of the leading strand.
Subunit
Homotrimer. Forms a complex with activator 1 heteropentamer in the presence of ATP (By similarity).
Homotrimer. Forms a complex with activator 1 heteropentamer in the presence of ATP (By similarity). Interacts with E2f.
Homotrimer. Forms a complex with activator 1 heteropentamer in the presence of ATP (By similarity). Interacts with E2f.
Similarity
Belongs to the PCNA family.
Keywords
Complete proteome
DNA replication
DNA-binding
Nucleus
Reference proteome
3D-structure
Direct protein sequencing
Feature
chain Proliferating cell nuclear antigen
Uniprot
H9JN03
Q8MYA5
Q8MYA4
O01377
S4PB42
A0A212ES54
+ More
A0A2A4J0R7 A0A194RJ19 I4DQ38 A0A2J7Q7K9 A0A067QR99 A0A1L8ECY7 A0A1J1I1Z1 A0A336KDE0 A0A2P8Y1H5 A0A1I8PWB0 A0A224XM68 A0A0P4VTM6 A0A023F9X0 R4G4N7 E9HC90 U5EWY5 A0A164ZBV7 A0A2Z5TRD7 T1P852 A0A0L0CN76 Q4PKD7 A0A023EMS3 R4V0H7 A0A182JD53 A0A084WD25 A0A0P6D7Z5 Q4PKD8 A0A0P5UH05 A0A182YHC7 A0A182T2U7 A0A182R6W2 A0A0P6EFX2 A0A0P5WMK4 A0A0N8C701 A0A1Q3EVV1 A0A0P5TGP9 A0A1B6GBS0 A0A1B6IC67 A0A0P5HVH6 A0A0P5DE32 A0A0P5MR45 A0A0N8CWA4 A0A0P4YU64 A0A0P5XF32 A0A0P6A769 G1E6N7 A0A1D8DDR6 A0A0P6HFC2 A0A182PV54 A0A182N8Q9 A0A182VSY1 A0A182UNS0 A0A182JPH1 A0A182TUZ9 A0A182KJS9 A0A182XB68 Q7Q0Q0 A0A182I0R2 W5JM12 A0A182FMV3 A0A1A9Y549 A0A1B0BXV0 A0A0P6EHN8 B5SP70 A0A1B0CN07 B1A9K0 A0A0P6DIL4 A0A1B6KVA0 A0A1L8DX45 A0A0P6DME1 A0A0P5P6P2 B0WPF0 A0A182QF77 O16852 A0A0P5P1J3 A0A1A9ZP02 A0A0P5NQD1 D3TQX0 A0A2A4J1Z5 W8B157 A0A1A9WKC2 A0A1A9VQ32 B4MRX3 B4KS28 A0A0K8W5Y1 A0A034WQG3 A0A3B0JJA1 Q28ZI4 B4GHQ3 A0A1W4V4B4 B3MHM6 B3NK47 P17917 A0A2H4T2M1
A0A2A4J0R7 A0A194RJ19 I4DQ38 A0A2J7Q7K9 A0A067QR99 A0A1L8ECY7 A0A1J1I1Z1 A0A336KDE0 A0A2P8Y1H5 A0A1I8PWB0 A0A224XM68 A0A0P4VTM6 A0A023F9X0 R4G4N7 E9HC90 U5EWY5 A0A164ZBV7 A0A2Z5TRD7 T1P852 A0A0L0CN76 Q4PKD7 A0A023EMS3 R4V0H7 A0A182JD53 A0A084WD25 A0A0P6D7Z5 Q4PKD8 A0A0P5UH05 A0A182YHC7 A0A182T2U7 A0A182R6W2 A0A0P6EFX2 A0A0P5WMK4 A0A0N8C701 A0A1Q3EVV1 A0A0P5TGP9 A0A1B6GBS0 A0A1B6IC67 A0A0P5HVH6 A0A0P5DE32 A0A0P5MR45 A0A0N8CWA4 A0A0P4YU64 A0A0P5XF32 A0A0P6A769 G1E6N7 A0A1D8DDR6 A0A0P6HFC2 A0A182PV54 A0A182N8Q9 A0A182VSY1 A0A182UNS0 A0A182JPH1 A0A182TUZ9 A0A182KJS9 A0A182XB68 Q7Q0Q0 A0A182I0R2 W5JM12 A0A182FMV3 A0A1A9Y549 A0A1B0BXV0 A0A0P6EHN8 B5SP70 A0A1B0CN07 B1A9K0 A0A0P6DIL4 A0A1B6KVA0 A0A1L8DX45 A0A0P6DME1 A0A0P5P6P2 B0WPF0 A0A182QF77 O16852 A0A0P5P1J3 A0A1A9ZP02 A0A0P5NQD1 D3TQX0 A0A2A4J1Z5 W8B157 A0A1A9WKC2 A0A1A9VQ32 B4MRX3 B4KS28 A0A0K8W5Y1 A0A034WQG3 A0A3B0JJA1 Q28ZI4 B4GHQ3 A0A1W4V4B4 B3MHM6 B3NK47 P17917 A0A2H4T2M1
Pubmed
19121390
9498568
23622113
22118469
26354079
22651552
+ More
24845553 29403074 27129103 25474469 21292972 26760975 25315136 26108605 16640730 17510324 24945155 26483478 24438588 25244985 24728082 20966253 12364791 14747013 17210077 20920257 23761445 18678269 20230291 9714841 20353571 24495485 17994087 25348373 15632085 1968224 10731132 12537572 12537569 1973166 7907981 9310333 19081076
24845553 29403074 27129103 25474469 21292972 26760975 25315136 26108605 16640730 17510324 24945155 26483478 24438588 25244985 24728082 20966253 12364791 14747013 17210077 20920257 23761445 18678269 20230291 9714841 20353571 24495485 17994087 25348373 15632085 1968224 10731132 12537572 12537569 1973166 7907981 9310333 19081076
EMBL
BABH01029174
AB069854
AB174843
BAC02929.1
BAD13299.1
AB069855
+ More
AB174894 BAC02930.1 BAD13316.1 AB002264 AB002265 GAIX01006122 JAA86438.1 AGBW02012888 OWR44281.1 NWSH01003854 PCG65757.1 KQ460323 KPJ15936.1 AK403915 BAM20028.1 NEVH01017442 PNF24568.1 KK853114 KDR11193.1 GFDG01002287 JAV16512.1 CVRI01000038 CRK94227.1 UFQS01000086 UFQT01000086 SSW99110.1 SSX19492.1 PYGN01001050 LT717627 PSN38108.1 SIW61619.1 GFTR01004242 JAW12184.1 GDKW01000724 JAI55871.1 GBBI01000517 JAC18195.1 ACPB03017886 GAHY01000616 JAA76894.1 GL732619 EFX70582.1 GANO01001211 JAB58660.1 LRGB01000745 KZS16181.1 FX985793 BBA93680.1 KA644757 AFP59386.1 JRES01000147 KNC33798.1 DQ075690 CH477894 AAY82461.1 EAT35273.1 JXUM01032592 GAPW01003453 KQ560961 JAC10145.1 KXJ80195.1 KC632288 AGM32102.1 ATLV01022923 KE525338 KFB48119.1 GDIQ01221864 GDIQ01095506 GDIQ01081966 GDIQ01073817 JAN12771.1 DQ075689 AAY82460.1 GDIP01113057 JAL90657.1 GDIQ01064324 GDIQ01064270 GDIQ01038938 JAN30467.1 GDIP01084101 JAM19614.1 GDIQ01108651 JAL43075.1 GFDL01015612 JAV19433.1 GDIP01132362 JAL71352.1 GECZ01010026 JAS59743.1 GECU01023180 JAS84526.1 GDIQ01224562 GDIQ01097258 GDIQ01095557 JAK27163.1 GDIP01158842 JAJ64560.1 GDIQ01153189 JAK98536.1 GDIP01092615 JAM11100.1 GDIP01223549 JAI99852.1 GDIP01073120 JAM30595.1 GDIP01034204 JAM69511.1 JN034913 JN034649 QCYY01002249 AEJ89927.1 AEJ89928.1 ROT71780.1 KU380336 AOS87743.1 GDIQ01020242 JAN74495.1 AAAB01008980 EAA13806.2 APCN01005234 ADMH02001180 ETN63819.1 JXJN01022373 GDIQ01076566 JAN18171.1 EF051247 ABM66815.1 AJWK01019585 EU431336 ACA09718.1 GDIQ01076568 JAN18169.1 GEBQ01024656 JAT15321.1 GFDF01003095 JAV10989.1 GDIQ01076567 JAN18170.1 GDIQ01139294 JAL12432.1 DS232023 EDS32329.1 AXCN02001859 AF020427 GDIQ01139296 JAL12430.1 GDIQ01139295 JAL12431.1 CCAG010006225 EZ423822 ADD20098.1 PCG65756.1 GAMC01014318 JAB92237.1 CH963850 EDW74862.1 CH933808 EDW10464.1 GDHF01005855 JAI46459.1 GAKP01002385 JAC56567.1 OUUW01000001 SPP73306.1 CM000071 EAL25629.1 CH479183 EDW36023.1 CH902619 EDV35862.1 CH954179 EDV55282.1 M33950 AE013599 AY122197 KY438972 ATY70175.1
AB174894 BAC02930.1 BAD13316.1 AB002264 AB002265 GAIX01006122 JAA86438.1 AGBW02012888 OWR44281.1 NWSH01003854 PCG65757.1 KQ460323 KPJ15936.1 AK403915 BAM20028.1 NEVH01017442 PNF24568.1 KK853114 KDR11193.1 GFDG01002287 JAV16512.1 CVRI01000038 CRK94227.1 UFQS01000086 UFQT01000086 SSW99110.1 SSX19492.1 PYGN01001050 LT717627 PSN38108.1 SIW61619.1 GFTR01004242 JAW12184.1 GDKW01000724 JAI55871.1 GBBI01000517 JAC18195.1 ACPB03017886 GAHY01000616 JAA76894.1 GL732619 EFX70582.1 GANO01001211 JAB58660.1 LRGB01000745 KZS16181.1 FX985793 BBA93680.1 KA644757 AFP59386.1 JRES01000147 KNC33798.1 DQ075690 CH477894 AAY82461.1 EAT35273.1 JXUM01032592 GAPW01003453 KQ560961 JAC10145.1 KXJ80195.1 KC632288 AGM32102.1 ATLV01022923 KE525338 KFB48119.1 GDIQ01221864 GDIQ01095506 GDIQ01081966 GDIQ01073817 JAN12771.1 DQ075689 AAY82460.1 GDIP01113057 JAL90657.1 GDIQ01064324 GDIQ01064270 GDIQ01038938 JAN30467.1 GDIP01084101 JAM19614.1 GDIQ01108651 JAL43075.1 GFDL01015612 JAV19433.1 GDIP01132362 JAL71352.1 GECZ01010026 JAS59743.1 GECU01023180 JAS84526.1 GDIQ01224562 GDIQ01097258 GDIQ01095557 JAK27163.1 GDIP01158842 JAJ64560.1 GDIQ01153189 JAK98536.1 GDIP01092615 JAM11100.1 GDIP01223549 JAI99852.1 GDIP01073120 JAM30595.1 GDIP01034204 JAM69511.1 JN034913 JN034649 QCYY01002249 AEJ89927.1 AEJ89928.1 ROT71780.1 KU380336 AOS87743.1 GDIQ01020242 JAN74495.1 AAAB01008980 EAA13806.2 APCN01005234 ADMH02001180 ETN63819.1 JXJN01022373 GDIQ01076566 JAN18171.1 EF051247 ABM66815.1 AJWK01019585 EU431336 ACA09718.1 GDIQ01076568 JAN18169.1 GEBQ01024656 JAT15321.1 GFDF01003095 JAV10989.1 GDIQ01076567 JAN18170.1 GDIQ01139294 JAL12432.1 DS232023 EDS32329.1 AXCN02001859 AF020427 GDIQ01139296 JAL12430.1 GDIQ01139295 JAL12431.1 CCAG010006225 EZ423822 ADD20098.1 PCG65756.1 GAMC01014318 JAB92237.1 CH963850 EDW74862.1 CH933808 EDW10464.1 GDHF01005855 JAI46459.1 GAKP01002385 JAC56567.1 OUUW01000001 SPP73306.1 CM000071 EAL25629.1 CH479183 EDW36023.1 CH902619 EDV35862.1 CH954179 EDV55282.1 M33950 AE013599 AY122197 KY438972 ATY70175.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000235965
UP000027135
+ More
UP000183832 UP000245037 UP000095300 UP000015103 UP000000305 UP000076858 UP000095301 UP000037069 UP000008820 UP000069940 UP000249989 UP000075880 UP000030765 UP000076408 UP000075901 UP000075900 UP000283509 UP000075885 UP000075884 UP000075920 UP000075903 UP000075881 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000000673 UP000069272 UP000092443 UP000092460 UP000092461 UP000002320 UP000075886 UP000092445 UP000092444 UP000091820 UP000078200 UP000007798 UP000009192 UP000268350 UP000001819 UP000008744 UP000192221 UP000007801 UP000008711 UP000000803
UP000183832 UP000245037 UP000095300 UP000015103 UP000000305 UP000076858 UP000095301 UP000037069 UP000008820 UP000069940 UP000249989 UP000075880 UP000030765 UP000076408 UP000075901 UP000075900 UP000283509 UP000075885 UP000075884 UP000075920 UP000075903 UP000075881 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000000673 UP000069272 UP000092443 UP000092460 UP000092461 UP000002320 UP000075886 UP000092445 UP000092444 UP000091820 UP000078200 UP000007798 UP000009192 UP000268350 UP000001819 UP000008744 UP000192221 UP000007801 UP000008711 UP000000803
Interpro
ProteinModelPortal
H9JN03
Q8MYA5
Q8MYA4
O01377
S4PB42
A0A212ES54
+ More
A0A2A4J0R7 A0A194RJ19 I4DQ38 A0A2J7Q7K9 A0A067QR99 A0A1L8ECY7 A0A1J1I1Z1 A0A336KDE0 A0A2P8Y1H5 A0A1I8PWB0 A0A224XM68 A0A0P4VTM6 A0A023F9X0 R4G4N7 E9HC90 U5EWY5 A0A164ZBV7 A0A2Z5TRD7 T1P852 A0A0L0CN76 Q4PKD7 A0A023EMS3 R4V0H7 A0A182JD53 A0A084WD25 A0A0P6D7Z5 Q4PKD8 A0A0P5UH05 A0A182YHC7 A0A182T2U7 A0A182R6W2 A0A0P6EFX2 A0A0P5WMK4 A0A0N8C701 A0A1Q3EVV1 A0A0P5TGP9 A0A1B6GBS0 A0A1B6IC67 A0A0P5HVH6 A0A0P5DE32 A0A0P5MR45 A0A0N8CWA4 A0A0P4YU64 A0A0P5XF32 A0A0P6A769 G1E6N7 A0A1D8DDR6 A0A0P6HFC2 A0A182PV54 A0A182N8Q9 A0A182VSY1 A0A182UNS0 A0A182JPH1 A0A182TUZ9 A0A182KJS9 A0A182XB68 Q7Q0Q0 A0A182I0R2 W5JM12 A0A182FMV3 A0A1A9Y549 A0A1B0BXV0 A0A0P6EHN8 B5SP70 A0A1B0CN07 B1A9K0 A0A0P6DIL4 A0A1B6KVA0 A0A1L8DX45 A0A0P6DME1 A0A0P5P6P2 B0WPF0 A0A182QF77 O16852 A0A0P5P1J3 A0A1A9ZP02 A0A0P5NQD1 D3TQX0 A0A2A4J1Z5 W8B157 A0A1A9WKC2 A0A1A9VQ32 B4MRX3 B4KS28 A0A0K8W5Y1 A0A034WQG3 A0A3B0JJA1 Q28ZI4 B4GHQ3 A0A1W4V4B4 B3MHM6 B3NK47 P17917 A0A2H4T2M1
A0A2A4J0R7 A0A194RJ19 I4DQ38 A0A2J7Q7K9 A0A067QR99 A0A1L8ECY7 A0A1J1I1Z1 A0A336KDE0 A0A2P8Y1H5 A0A1I8PWB0 A0A224XM68 A0A0P4VTM6 A0A023F9X0 R4G4N7 E9HC90 U5EWY5 A0A164ZBV7 A0A2Z5TRD7 T1P852 A0A0L0CN76 Q4PKD7 A0A023EMS3 R4V0H7 A0A182JD53 A0A084WD25 A0A0P6D7Z5 Q4PKD8 A0A0P5UH05 A0A182YHC7 A0A182T2U7 A0A182R6W2 A0A0P6EFX2 A0A0P5WMK4 A0A0N8C701 A0A1Q3EVV1 A0A0P5TGP9 A0A1B6GBS0 A0A1B6IC67 A0A0P5HVH6 A0A0P5DE32 A0A0P5MR45 A0A0N8CWA4 A0A0P4YU64 A0A0P5XF32 A0A0P6A769 G1E6N7 A0A1D8DDR6 A0A0P6HFC2 A0A182PV54 A0A182N8Q9 A0A182VSY1 A0A182UNS0 A0A182JPH1 A0A182TUZ9 A0A182KJS9 A0A182XB68 Q7Q0Q0 A0A182I0R2 W5JM12 A0A182FMV3 A0A1A9Y549 A0A1B0BXV0 A0A0P6EHN8 B5SP70 A0A1B0CN07 B1A9K0 A0A0P6DIL4 A0A1B6KVA0 A0A1L8DX45 A0A0P6DME1 A0A0P5P6P2 B0WPF0 A0A182QF77 O16852 A0A0P5P1J3 A0A1A9ZP02 A0A0P5NQD1 D3TQX0 A0A2A4J1Z5 W8B157 A0A1A9WKC2 A0A1A9VQ32 B4MRX3 B4KS28 A0A0K8W5Y1 A0A034WQG3 A0A3B0JJA1 Q28ZI4 B4GHQ3 A0A1W4V4B4 B3MHM6 B3NK47 P17917 A0A2H4T2M1
PDB
4CS5
E-value=1.19049e-112,
Score=1037
Ontologies
KEGG
PATHWAY
GO
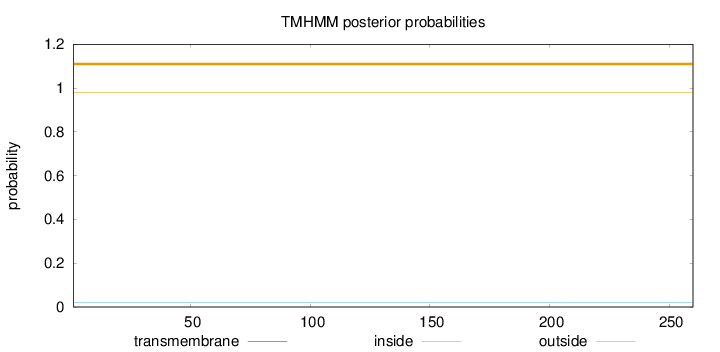
Topology
Subcellular location
Nucleus
Length:
260
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00032
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01924
outside
1 - 260
Population Genetic Test Statistics
Pi
19.370683
Theta
18.682668
Tajima's D
-0.256169
CLR
1.152863
CSRT
0.305184740762962
Interpretation
Uncertain
