Gene
KWMTBOMO13401
Annotation
PREDICTED:_uncharacterized_protein_LOC106709836_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 1.702 Mitochondrial Reliability : 1.012 Nuclear Reliability : 1.232
Sequence
CDS
ATGTTAACGTTTTGTTTATCAGATCGCCACACGCATTCGCCTGAAGTGGAACGTGAGCGAGCTACATTAACGGAGCTGTTGTCTACAATGACCCTGGGACAACTGTTACCGCATCCGCTGAGCCATATGTTGGAATTAGCACCTGAGCTTACTCCGAACGAGATTGTACAAGTACTCCGTGACTGTTTGTGGAACTACACACGGGACCACGTCCCGTCGCCTGCTTTATTTACGTGTGACGCTACAGGCTTGCACTGGCGCGATCCGTCATCTTGCAAACCACCGGCCTTATATACGGATACTTTGAGGATTATAATACAGAAAAGAATAGCGAAATTGGGACATTTGTATCCCATAATGTTTTTGAATCTTGAAAAGGAAAGCAGTAATTAA
Protein
MLTFCLSDRHTHSPEVERERATLTELLSTMTLGQLLPHPLSHMLELAPELTPNEIVQVLRDCLWNYTRDHVPSPALFTCDATGLHWRDPSSCKPPALYTDTLRIIIQKRIAKLGHLYPIMFLNLEKESSN
Summary
Uniprot
A0A194PXL7
A0A194RDL5
A0A212ES16
A0A1Y1KY82
A0A0T6AUL1
A0A1Y1KVU4
+ More
A0A1B6D092 A0A1B6DCC8 D6X1X5 A0A1B6HAM8 N6U9Y1 U4UJ96 A0A1B6EJY2 A0A1B6JTM8 A0A1Q3EZY9 A0A1W4X5W2 A0A1W4WWF7 A0A1W4X798 A0A0A9YRL2 A0A1B6LJB1 A0A1B6H8L2 A0A067R9K9 A0A1B6FNZ4 A0A2P8Y6E3 A0A146LYA2 A0A2J7QVU0 A0A084WTF4 A0A182IPK9 A0A0K8SYI2 A0A1B0DPW1 A0A0A9YL05 A0A182FR89 W5JF79 A0A182RPM8 A0A182LTV3 A0A182VQ75 A0A1B0CEH9 A0A182P578 A0A182TMB7 Q7QBI6 A0A182UMI5 A0A182XE75 A0A182HJ48 A0A182LC99 A0A182Y7B5 T1HYQ9 A0A182SH31 A0A182N4L2 A0A1S3D106 E0VWT3 A0A182QUE0 J9K396 A0A336LS77 A0A023EZ71 A0A224XJD9 A0A2S2PRX7 A0A1A9V754 A0A2H8TD34 A0A2R7WN93 A0A0M5JCW4 A0A0M4ERW6 A0A1A9Z490 A0A1B0BJZ1 A0A1A9XJ13 A0A1B0FGM7 A0A1A9W609 W8B1T7 A0A034WLK3 A0A0L0CE29 A0A0K8VUU4 A0A1I8PX62 T1PHE5 A0A034WJZ0 A0A0A1XRW4 A0A2S2QAP0 A0A1I8MJP2 E2B700 B4KA51 B4M0X2 A0A026WQ17 A0A195BA51 A0A158NK46 F4W8D4 A0A195FAF5 A0A151X293 A0A3B0K0J7 B4G2U2 Q297P5 B4JI97 A0A1W4V669 A0A151JME2 A0A195C2U6 A0A310SDQ2 T1GJ15 B3LXB0 A0A0C9QYZ4
A0A1B6D092 A0A1B6DCC8 D6X1X5 A0A1B6HAM8 N6U9Y1 U4UJ96 A0A1B6EJY2 A0A1B6JTM8 A0A1Q3EZY9 A0A1W4X5W2 A0A1W4WWF7 A0A1W4X798 A0A0A9YRL2 A0A1B6LJB1 A0A1B6H8L2 A0A067R9K9 A0A1B6FNZ4 A0A2P8Y6E3 A0A146LYA2 A0A2J7QVU0 A0A084WTF4 A0A182IPK9 A0A0K8SYI2 A0A1B0DPW1 A0A0A9YL05 A0A182FR89 W5JF79 A0A182RPM8 A0A182LTV3 A0A182VQ75 A0A1B0CEH9 A0A182P578 A0A182TMB7 Q7QBI6 A0A182UMI5 A0A182XE75 A0A182HJ48 A0A182LC99 A0A182Y7B5 T1HYQ9 A0A182SH31 A0A182N4L2 A0A1S3D106 E0VWT3 A0A182QUE0 J9K396 A0A336LS77 A0A023EZ71 A0A224XJD9 A0A2S2PRX7 A0A1A9V754 A0A2H8TD34 A0A2R7WN93 A0A0M5JCW4 A0A0M4ERW6 A0A1A9Z490 A0A1B0BJZ1 A0A1A9XJ13 A0A1B0FGM7 A0A1A9W609 W8B1T7 A0A034WLK3 A0A0L0CE29 A0A0K8VUU4 A0A1I8PX62 T1PHE5 A0A034WJZ0 A0A0A1XRW4 A0A2S2QAP0 A0A1I8MJP2 E2B700 B4KA51 B4M0X2 A0A026WQ17 A0A195BA51 A0A158NK46 F4W8D4 A0A195FAF5 A0A151X293 A0A3B0K0J7 B4G2U2 Q297P5 B4JI97 A0A1W4V669 A0A151JME2 A0A195C2U6 A0A310SDQ2 T1GJ15 B3LXB0 A0A0C9QYZ4
Pubmed
26354079
22118469
28004739
18362917
19820115
23537049
+ More
25401762 24845553 29403074 26823975 24438588 20920257 23761445 12364791 14747013 17210077 20966253 25244985 20566863 25474469 24495485 25348373 26108605 25830018 25315136 20798317 17994087 24508170 30249741 21347285 21719571 15632085 23185243
25401762 24845553 29403074 26823975 24438588 20920257 23761445 12364791 14747013 17210077 20966253 25244985 20566863 25474469 24495485 25348373 26108605 25830018 25315136 20798317 17994087 24508170 30249741 21347285 21719571 15632085 23185243
EMBL
KQ459586
KPI98067.1
KQ460323
KPJ15933.1
AGBW02012888
OWR44283.1
+ More
GEZM01072446 JAV65518.1 LJIG01022844 KRT78423.1 GEZM01072445 JAV65519.1 GEDC01018191 JAS19107.1 GEDC01013980 JAS23318.1 KQ971371 EFA10179.2 GECU01036006 JAS71700.1 APGK01043340 KB741014 ENN75427.1 KB632381 ERL94154.1 GECZ01031520 JAS38249.1 GECU01005128 JAT02579.1 GFDL01014178 JAV20867.1 GBHO01009861 JAG33743.1 GEBQ01016278 JAT23699.1 GECU01036743 JAS70963.1 KK852801 KDR16305.1 GECZ01017895 JAS51874.1 PYGN01000872 PSN39828.1 GDHC01006031 JAQ12598.1 NEVH01009770 PNF32705.1 ATLV01026890 KE525420 KFB53498.1 GBRD01007478 JAG58343.1 AJVK01018647 AJVK01018648 GBHO01009862 JAG33742.1 ADMH02001656 ETN61540.1 AXCM01000623 AJWK01008878 AAAB01008879 EAA08442.5 APCN01002124 ACPB03001414 DS235824 EEB17839.1 AXCN02000045 ABLF02019319 UFQS01000141 UFQT01000141 SSX00400.1 SSX20780.1 GBBI01004666 JAC14046.1 GFTR01008285 JAW08141.1 GGMR01019610 MBY32229.1 GFXV01000195 MBW12000.1 KK855137 PTY21092.1 CP012526 ALC47356.1 ALC45267.1 JXJN01015757 CCAG010002945 GAMC01015582 JAB90973.1 GAKP01004304 JAC54648.1 JRES01000501 KNC30668.1 GDHF01010009 JAI42305.1 KA648247 AFP62876.1 GAKP01004305 JAC54647.1 GBXI01000575 JAD13717.1 GGMS01005616 MBY74819.1 GL446071 EFN88542.1 CH933806 EDW16726.1 CH940650 EDW67383.1 KK107133 QOIP01000011 EZA58122.1 RLU16460.1 KQ976537 KYM81411.1 ADTU01018656 ADTU01018657 GL887908 EGI69425.1 KQ981693 KYN37605.1 KQ982579 KYQ54555.1 OUUW01000013 SPP87835.1 CH479179 EDW24137.1 CM000070 EAL28160.4 KRT00509.1 CH916369 EDV92978.1 KQ978928 KYN27489.1 KQ978344 KYM94915.1 KQ760974 OAD58560.1 CAQQ02191425 CH902617 EDV41710.1 GBYB01005902 JAG75669.1
GEZM01072446 JAV65518.1 LJIG01022844 KRT78423.1 GEZM01072445 JAV65519.1 GEDC01018191 JAS19107.1 GEDC01013980 JAS23318.1 KQ971371 EFA10179.2 GECU01036006 JAS71700.1 APGK01043340 KB741014 ENN75427.1 KB632381 ERL94154.1 GECZ01031520 JAS38249.1 GECU01005128 JAT02579.1 GFDL01014178 JAV20867.1 GBHO01009861 JAG33743.1 GEBQ01016278 JAT23699.1 GECU01036743 JAS70963.1 KK852801 KDR16305.1 GECZ01017895 JAS51874.1 PYGN01000872 PSN39828.1 GDHC01006031 JAQ12598.1 NEVH01009770 PNF32705.1 ATLV01026890 KE525420 KFB53498.1 GBRD01007478 JAG58343.1 AJVK01018647 AJVK01018648 GBHO01009862 JAG33742.1 ADMH02001656 ETN61540.1 AXCM01000623 AJWK01008878 AAAB01008879 EAA08442.5 APCN01002124 ACPB03001414 DS235824 EEB17839.1 AXCN02000045 ABLF02019319 UFQS01000141 UFQT01000141 SSX00400.1 SSX20780.1 GBBI01004666 JAC14046.1 GFTR01008285 JAW08141.1 GGMR01019610 MBY32229.1 GFXV01000195 MBW12000.1 KK855137 PTY21092.1 CP012526 ALC47356.1 ALC45267.1 JXJN01015757 CCAG010002945 GAMC01015582 JAB90973.1 GAKP01004304 JAC54648.1 JRES01000501 KNC30668.1 GDHF01010009 JAI42305.1 KA648247 AFP62876.1 GAKP01004305 JAC54647.1 GBXI01000575 JAD13717.1 GGMS01005616 MBY74819.1 GL446071 EFN88542.1 CH933806 EDW16726.1 CH940650 EDW67383.1 KK107133 QOIP01000011 EZA58122.1 RLU16460.1 KQ976537 KYM81411.1 ADTU01018656 ADTU01018657 GL887908 EGI69425.1 KQ981693 KYN37605.1 KQ982579 KYQ54555.1 OUUW01000013 SPP87835.1 CH479179 EDW24137.1 CM000070 EAL28160.4 KRT00509.1 CH916369 EDV92978.1 KQ978928 KYN27489.1 KQ978344 KYM94915.1 KQ760974 OAD58560.1 CAQQ02191425 CH902617 EDV41710.1 GBYB01005902 JAG75669.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000007266
UP000019118
UP000030742
+ More
UP000192223 UP000027135 UP000245037 UP000235965 UP000030765 UP000075880 UP000092462 UP000069272 UP000000673 UP000075900 UP000075883 UP000075920 UP000092461 UP000075885 UP000075902 UP000007062 UP000075903 UP000076407 UP000075840 UP000075882 UP000076408 UP000015103 UP000075901 UP000075884 UP000079169 UP000009046 UP000075886 UP000007819 UP000078200 UP000092553 UP000092445 UP000092460 UP000092443 UP000092444 UP000091820 UP000037069 UP000095300 UP000095301 UP000008237 UP000009192 UP000008792 UP000053097 UP000279307 UP000078540 UP000005205 UP000007755 UP000078541 UP000075809 UP000268350 UP000008744 UP000001819 UP000001070 UP000192221 UP000078492 UP000078542 UP000015102 UP000007801
UP000192223 UP000027135 UP000245037 UP000235965 UP000030765 UP000075880 UP000092462 UP000069272 UP000000673 UP000075900 UP000075883 UP000075920 UP000092461 UP000075885 UP000075902 UP000007062 UP000075903 UP000076407 UP000075840 UP000075882 UP000076408 UP000015103 UP000075901 UP000075884 UP000079169 UP000009046 UP000075886 UP000007819 UP000078200 UP000092553 UP000092445 UP000092460 UP000092443 UP000092444 UP000091820 UP000037069 UP000095300 UP000095301 UP000008237 UP000009192 UP000008792 UP000053097 UP000279307 UP000078540 UP000005205 UP000007755 UP000078541 UP000075809 UP000268350 UP000008744 UP000001819 UP000001070 UP000192221 UP000078492 UP000078542 UP000015102 UP000007801
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
A0A194PXL7
A0A194RDL5
A0A212ES16
A0A1Y1KY82
A0A0T6AUL1
A0A1Y1KVU4
+ More
A0A1B6D092 A0A1B6DCC8 D6X1X5 A0A1B6HAM8 N6U9Y1 U4UJ96 A0A1B6EJY2 A0A1B6JTM8 A0A1Q3EZY9 A0A1W4X5W2 A0A1W4WWF7 A0A1W4X798 A0A0A9YRL2 A0A1B6LJB1 A0A1B6H8L2 A0A067R9K9 A0A1B6FNZ4 A0A2P8Y6E3 A0A146LYA2 A0A2J7QVU0 A0A084WTF4 A0A182IPK9 A0A0K8SYI2 A0A1B0DPW1 A0A0A9YL05 A0A182FR89 W5JF79 A0A182RPM8 A0A182LTV3 A0A182VQ75 A0A1B0CEH9 A0A182P578 A0A182TMB7 Q7QBI6 A0A182UMI5 A0A182XE75 A0A182HJ48 A0A182LC99 A0A182Y7B5 T1HYQ9 A0A182SH31 A0A182N4L2 A0A1S3D106 E0VWT3 A0A182QUE0 J9K396 A0A336LS77 A0A023EZ71 A0A224XJD9 A0A2S2PRX7 A0A1A9V754 A0A2H8TD34 A0A2R7WN93 A0A0M5JCW4 A0A0M4ERW6 A0A1A9Z490 A0A1B0BJZ1 A0A1A9XJ13 A0A1B0FGM7 A0A1A9W609 W8B1T7 A0A034WLK3 A0A0L0CE29 A0A0K8VUU4 A0A1I8PX62 T1PHE5 A0A034WJZ0 A0A0A1XRW4 A0A2S2QAP0 A0A1I8MJP2 E2B700 B4KA51 B4M0X2 A0A026WQ17 A0A195BA51 A0A158NK46 F4W8D4 A0A195FAF5 A0A151X293 A0A3B0K0J7 B4G2U2 Q297P5 B4JI97 A0A1W4V669 A0A151JME2 A0A195C2U6 A0A310SDQ2 T1GJ15 B3LXB0 A0A0C9QYZ4
A0A1B6D092 A0A1B6DCC8 D6X1X5 A0A1B6HAM8 N6U9Y1 U4UJ96 A0A1B6EJY2 A0A1B6JTM8 A0A1Q3EZY9 A0A1W4X5W2 A0A1W4WWF7 A0A1W4X798 A0A0A9YRL2 A0A1B6LJB1 A0A1B6H8L2 A0A067R9K9 A0A1B6FNZ4 A0A2P8Y6E3 A0A146LYA2 A0A2J7QVU0 A0A084WTF4 A0A182IPK9 A0A0K8SYI2 A0A1B0DPW1 A0A0A9YL05 A0A182FR89 W5JF79 A0A182RPM8 A0A182LTV3 A0A182VQ75 A0A1B0CEH9 A0A182P578 A0A182TMB7 Q7QBI6 A0A182UMI5 A0A182XE75 A0A182HJ48 A0A182LC99 A0A182Y7B5 T1HYQ9 A0A182SH31 A0A182N4L2 A0A1S3D106 E0VWT3 A0A182QUE0 J9K396 A0A336LS77 A0A023EZ71 A0A224XJD9 A0A2S2PRX7 A0A1A9V754 A0A2H8TD34 A0A2R7WN93 A0A0M5JCW4 A0A0M4ERW6 A0A1A9Z490 A0A1B0BJZ1 A0A1A9XJ13 A0A1B0FGM7 A0A1A9W609 W8B1T7 A0A034WLK3 A0A0L0CE29 A0A0K8VUU4 A0A1I8PX62 T1PHE5 A0A034WJZ0 A0A0A1XRW4 A0A2S2QAP0 A0A1I8MJP2 E2B700 B4KA51 B4M0X2 A0A026WQ17 A0A195BA51 A0A158NK46 F4W8D4 A0A195FAF5 A0A151X293 A0A3B0K0J7 B4G2U2 Q297P5 B4JI97 A0A1W4V669 A0A151JME2 A0A195C2U6 A0A310SDQ2 T1GJ15 B3LXB0 A0A0C9QYZ4
Ontologies
PANTHER
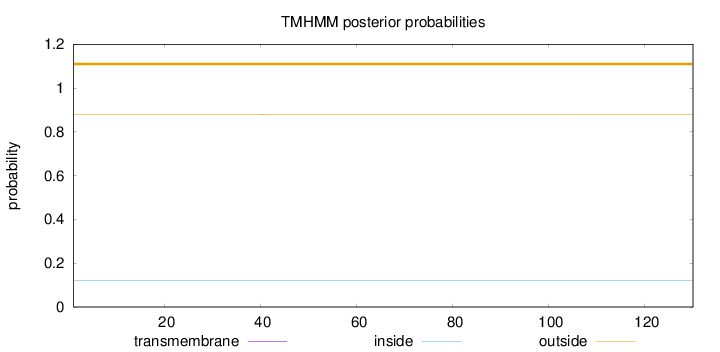
Topology
Length:
130
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00526
Exp number, first 60 AAs:
0.00343
Total prob of N-in:
0.12128
outside
1 - 130
Population Genetic Test Statistics
Pi
16.577075
Theta
13.714514
Tajima's D
-0.403006
CLR
0.486782
CSRT
0.262136893155342
Interpretation
Uncertain
