Gene
KWMTBOMO13400
Annotation
PREDICTED:_GPN-loop_GTPase_1-like_[Bombyx_mori]
Full name
GPN-loop GTPase
Location in the cell
Cytoplasmic Reliability : 2.114 Nuclear Reliability : 1.582
Sequence
CDS
ATGTCTTCCGATACAAATAAACCAGAAGGTTCAAACGCGGAGCCCAACAAACCCATTTGTTTGATAATTTTAGGTATGGCCGGCGCTGGAAAGACTTCCTTCACACGTCGATTGTCAAATAAAGTTATTAATGGATCACGACCTTATCTAGTGAACTTGGATCCAGCTTGCAGGGAAGTACCTTATCCTGCCAACATTGATATAAGAGATACTGTAGACTATAAAGAAGTAATGAAGCAGTATGGTCTTGGACCAAATGGTGGTATAGTTACAGCACTGAATCTGTTTTCAACAAAATTTGGACAAGTCGTAGACTTGATAGAGAAAGCGGGAAAGAAACACAAGTACTGTGTTATCGATACGCCAGGTCAAATAGAAGTGTTCACATGGTCAGCATCAGGCACAATAGTGACAGAAACACTGGCGTCATGCTGTCCCACAGTCGTAGTGTATGTAATGGACACAGTACGCAGTGTGTCGCCCGTCACATTTATGTCCAACATGCTGTATGCTTGCTCAATATTGTACAAGACTCGGCTGCCGTTTATTGTTGTCATGAATAAAATCGATGTAGTAAACAATTCATACGCAGTGGAATGGATGAGAGATTTCGAAGCTTTCCAAGAAGCATTAAATGCAGATGGTGGGGGAGAGACGGAAGGAGGAAATACTTACATAGGAAACTTGACACAGTCCATGGCACTGGCGTTGGACAGTTTTTATTCAGATCTGAAGTGCTGTGGTGTGAGTGCGCATACCGGTGAAGGCATAGATGAGTTCTTCAAACTAGTTGATGAATCTGCAGAGGAATATGAAAGAGAATACAAAGCGGATTGGTTGAAAATGCGCGCTGAAAAATTGGCAGAAGAGAAACGAAGGAAAGAAGATGAATTAAAAAAGACAGACAGCGTGGAGACAGTTATAGACAAGAAGAGTTTGATAGATGAAGTGCCGATCGGCAGAGAGCTCAGCGACATGTACCTGAAGCACCCCGGCAACGAGAGCTCCAGCGACGACGAGGGAACTGAAAATGAACCGGAAATCGACGACAAACCAGAGGACGTGGCTATGTTCCAAGAGTTCTTAAAGAAACACATAAAACCTAATGACACGCAGAAATAA
Protein
MSSDTNKPEGSNAEPNKPICLIILGMAGAGKTSFTRRLSNKVINGSRPYLVNLDPACREVPYPANIDIRDTVDYKEVMKQYGLGPNGGIVTALNLFSTKFGQVVDLIEKAGKKHKYCVIDTPGQIEVFTWSASGTIVTETLASCCPTVVVYVMDTVRSVSPVTFMSNMLYACSILYKTRLPFIVVMNKIDVVNNSYAVEWMRDFEAFQEALNADGGGETEGGNTYIGNLTQSMALALDSFYSDLKCCGVSAHTGEGIDEFFKLVDESAEEYEREYKADWLKMRAEKLAEEKRRKEDELKKTDSVETVIDKKSLIDEVPIGRELSDMYLKHPGNESSSDDEGTENEPEIDDKPEDVAMFQEFLKKHIKPNDTQK
Summary
Description
Small GTPase required for proper nuclear import of RNA polymerase II (RNAPII). May act at an RNAP assembly step prior to nuclear import.
Subunit
Binds to RNA polymerase II.
Similarity
Belongs to the GPN-loop GTPase family.
Uniprot
A0A2H1V049
A0A194REB6
A0A194PYG7
A0A3S2L685
A0A212ES60
A0A1E1W6Y9
+ More
S4PKV9 Q16TZ7 B0W660 A0A1Q3F4C3 A0A1Q3F4A8 A0A182GD34 A0A0M8ZXM4 A0A232EX19 K7IRL7 A0A154NXX6 A0A195F0K8 A0A0C9RCV1 A0A1J1HH57 E1ZX65 E9IEA5 A0A0T6B659 A0A195BYY8 A0A158P3K0 A0A1I8M785 A0A1B6CA99 A0A088ANI7 A0A067R7P6 A0A0L7R128 A0A1B6G7E8 A0A2A3EHW7 U5EIA7 A0A2J7R8A3 A0A0J7L9Q7 E2BLZ3 A0A182PNV1 A0A151IH85 A0A182YDY3 A0A151IV51 A0A1B6IYN3 A0A1A9W8R4 A0A1B0DQV1 A0A1A9USZ5 A0A182FLY6 A0A182QS09 A0A1B0FHM2 A0A182NP70 A0A2P8Y1P6 A0A182G371 A0A1A9Z6C1 A0A1L8DWV2 A0A151XDL8 A0A1A9XNK6 F4WAT3 A0A182RWX9 A0A084VJI3 A0A0K8TTP7 A0A1I8NY79 A0A1B0BNV4 W5JCN2 A0A2M4AM18 A0A182W7X2 A0A182LYH5 A0A2M4BRB5 A0A182J789 A0A1B0CFG7 A0A1W4X8C9 A0A182SLD7 A0A1W4X747 A0A0K8W8M5 A0A0L0BZ32 W8CCV9 D2A4G3 A0A2M4BS91 A0A2M4BSI3 A0A2R7VTM9 A0A3B5AIY4 A0A1W5ACE9 A0A1W5AKN7 A0A0A1WTT9 A0A1S3RAU2 A0A0S7LWP7 A0A210PG82 A0A3B3Y372 A0A3Q0JGC9 A0A3B3W2H1 A0A087X2N6 G3Q0C1 A0A3P8RL57 A0A2D0RMY4 A0A315VV76 A0A1Y1NK86 G3Q0C4 A0A3Q2X6R5 A0A3B4TU02 A0A147ARZ8 A0A3Q1IXY4 A0A3S2N614
S4PKV9 Q16TZ7 B0W660 A0A1Q3F4C3 A0A1Q3F4A8 A0A182GD34 A0A0M8ZXM4 A0A232EX19 K7IRL7 A0A154NXX6 A0A195F0K8 A0A0C9RCV1 A0A1J1HH57 E1ZX65 E9IEA5 A0A0T6B659 A0A195BYY8 A0A158P3K0 A0A1I8M785 A0A1B6CA99 A0A088ANI7 A0A067R7P6 A0A0L7R128 A0A1B6G7E8 A0A2A3EHW7 U5EIA7 A0A2J7R8A3 A0A0J7L9Q7 E2BLZ3 A0A182PNV1 A0A151IH85 A0A182YDY3 A0A151IV51 A0A1B6IYN3 A0A1A9W8R4 A0A1B0DQV1 A0A1A9USZ5 A0A182FLY6 A0A182QS09 A0A1B0FHM2 A0A182NP70 A0A2P8Y1P6 A0A182G371 A0A1A9Z6C1 A0A1L8DWV2 A0A151XDL8 A0A1A9XNK6 F4WAT3 A0A182RWX9 A0A084VJI3 A0A0K8TTP7 A0A1I8NY79 A0A1B0BNV4 W5JCN2 A0A2M4AM18 A0A182W7X2 A0A182LYH5 A0A2M4BRB5 A0A182J789 A0A1B0CFG7 A0A1W4X8C9 A0A182SLD7 A0A1W4X747 A0A0K8W8M5 A0A0L0BZ32 W8CCV9 D2A4G3 A0A2M4BS91 A0A2M4BSI3 A0A2R7VTM9 A0A3B5AIY4 A0A1W5ACE9 A0A1W5AKN7 A0A0A1WTT9 A0A1S3RAU2 A0A0S7LWP7 A0A210PG82 A0A3B3Y372 A0A3Q0JGC9 A0A3B3W2H1 A0A087X2N6 G3Q0C1 A0A3P8RL57 A0A2D0RMY4 A0A315VV76 A0A1Y1NK86 G3Q0C4 A0A3Q2X6R5 A0A3B4TU02 A0A147ARZ8 A0A3Q1IXY4 A0A3S2N614
EC Number
3.6.5.-
Pubmed
EMBL
ODYU01000073
SOQ34215.1
KQ460323
KPJ15932.1
KQ459586
KPI98068.1
+ More
RSAL01000124 RVE46613.1 AGBW02012888 OWR44284.1 GDQN01008337 JAT82717.1 GAIX01004320 JAA88240.1 CH477636 EAT37997.1 DS231847 EDS36407.1 GFDL01012676 JAV22369.1 GFDL01012682 JAV22363.1 JXUM01160707 KQ573791 KXJ67903.1 KQ435833 KOX71673.1 NNAY01001793 OXU22906.1 KQ434782 KZC04526.1 KQ981880 KYN33896.1 GBYB01006120 JAG75887.1 CVRI01000002 CRK86890.1 GL435030 EFN74155.1 GL762576 EFZ21091.1 LJIG01009583 KRT82790.1 KQ976394 KYM93136.1 ADTU01008166 GEDC01026944 GEDC01011887 JAS10354.1 JAS25411.1 KK852685 KDR18496.1 KQ414668 KOC64553.1 GECZ01011438 JAS58331.1 KZ288254 PBC30766.1 GANO01002704 JAB57167.1 NEVH01006723 PNF37073.1 LBMM01000143 KMR04775.1 GL449088 EFN83279.1 KQ977636 KYN01141.1 KQ980924 KYN11410.1 GECU01021099 GECU01019155 GECU01015687 JAS86607.1 JAS88551.1 JAS92019.1 AJVK01019382 AJVK01019383 AJVK01019384 AJVK01019385 AXCN02002424 CCAG010014882 PYGN01001050 PSN38114.1 JXUM01140972 JXUM01140973 KQ569162 KXJ68744.1 GFDF01003188 JAV10896.1 KQ982268 KYQ58476.1 GL888053 EGI68689.1 ATLV01013581 KE524868 KFB38127.1 GDAI01000283 JAI17320.1 JXJN01017617 ADMH02001606 ETN61841.1 GGFK01008461 MBW41782.1 AXCM01000973 GGFJ01006403 MBW55544.1 AXCP01007318 AJWK01010101 GDHF01004940 JAI47374.1 JRES01001224 KNC24494.1 GAMC01003016 JAC03540.1 KQ971344 EFA05223.1 GGFJ01006814 MBW55955.1 GGFJ01006813 MBW55954.1 KK854082 PTY10882.1 GBXI01011803 JAD02489.1 GBYX01141985 JAO93910.1 NEDP02076724 OWF35492.1 AYCK01020792 NHOQ01001616 PWA23321.1 GEZM01001136 JAV98129.1 GCES01005116 JAR81207.1 CM012439 RVE74919.1
RSAL01000124 RVE46613.1 AGBW02012888 OWR44284.1 GDQN01008337 JAT82717.1 GAIX01004320 JAA88240.1 CH477636 EAT37997.1 DS231847 EDS36407.1 GFDL01012676 JAV22369.1 GFDL01012682 JAV22363.1 JXUM01160707 KQ573791 KXJ67903.1 KQ435833 KOX71673.1 NNAY01001793 OXU22906.1 KQ434782 KZC04526.1 KQ981880 KYN33896.1 GBYB01006120 JAG75887.1 CVRI01000002 CRK86890.1 GL435030 EFN74155.1 GL762576 EFZ21091.1 LJIG01009583 KRT82790.1 KQ976394 KYM93136.1 ADTU01008166 GEDC01026944 GEDC01011887 JAS10354.1 JAS25411.1 KK852685 KDR18496.1 KQ414668 KOC64553.1 GECZ01011438 JAS58331.1 KZ288254 PBC30766.1 GANO01002704 JAB57167.1 NEVH01006723 PNF37073.1 LBMM01000143 KMR04775.1 GL449088 EFN83279.1 KQ977636 KYN01141.1 KQ980924 KYN11410.1 GECU01021099 GECU01019155 GECU01015687 JAS86607.1 JAS88551.1 JAS92019.1 AJVK01019382 AJVK01019383 AJVK01019384 AJVK01019385 AXCN02002424 CCAG010014882 PYGN01001050 PSN38114.1 JXUM01140972 JXUM01140973 KQ569162 KXJ68744.1 GFDF01003188 JAV10896.1 KQ982268 KYQ58476.1 GL888053 EGI68689.1 ATLV01013581 KE524868 KFB38127.1 GDAI01000283 JAI17320.1 JXJN01017617 ADMH02001606 ETN61841.1 GGFK01008461 MBW41782.1 AXCM01000973 GGFJ01006403 MBW55544.1 AXCP01007318 AJWK01010101 GDHF01004940 JAI47374.1 JRES01001224 KNC24494.1 GAMC01003016 JAC03540.1 KQ971344 EFA05223.1 GGFJ01006814 MBW55955.1 GGFJ01006813 MBW55954.1 KK854082 PTY10882.1 GBXI01011803 JAD02489.1 GBYX01141985 JAO93910.1 NEDP02076724 OWF35492.1 AYCK01020792 NHOQ01001616 PWA23321.1 GEZM01001136 JAV98129.1 GCES01005116 JAR81207.1 CM012439 RVE74919.1
Proteomes
UP000053240
UP000053268
UP000283053
UP000007151
UP000008820
UP000002320
+ More
UP000069940 UP000249989 UP000053105 UP000215335 UP000002358 UP000076502 UP000078541 UP000183832 UP000000311 UP000078540 UP000005205 UP000095301 UP000005203 UP000027135 UP000053825 UP000242457 UP000235965 UP000036403 UP000008237 UP000075885 UP000078542 UP000076408 UP000078492 UP000091820 UP000092462 UP000078200 UP000069272 UP000075886 UP000092444 UP000075884 UP000245037 UP000092445 UP000075809 UP000092443 UP000007755 UP000075900 UP000030765 UP000095300 UP000092460 UP000000673 UP000075920 UP000075883 UP000075880 UP000092461 UP000192223 UP000075901 UP000037069 UP000007266 UP000261400 UP000192224 UP000087266 UP000242188 UP000261480 UP000079169 UP000261500 UP000028760 UP000007635 UP000265080 UP000221080 UP000264840 UP000261420 UP000265040
UP000069940 UP000249989 UP000053105 UP000215335 UP000002358 UP000076502 UP000078541 UP000183832 UP000000311 UP000078540 UP000005205 UP000095301 UP000005203 UP000027135 UP000053825 UP000242457 UP000235965 UP000036403 UP000008237 UP000075885 UP000078542 UP000076408 UP000078492 UP000091820 UP000092462 UP000078200 UP000069272 UP000075886 UP000092444 UP000075884 UP000245037 UP000092445 UP000075809 UP000092443 UP000007755 UP000075900 UP000030765 UP000095300 UP000092460 UP000000673 UP000075920 UP000075883 UP000075880 UP000092461 UP000192223 UP000075901 UP000037069 UP000007266 UP000261400 UP000192224 UP000087266 UP000242188 UP000261480 UP000079169 UP000261500 UP000028760 UP000007635 UP000265080 UP000221080 UP000264840 UP000261420 UP000265040
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2H1V049
A0A194REB6
A0A194PYG7
A0A3S2L685
A0A212ES60
A0A1E1W6Y9
+ More
S4PKV9 Q16TZ7 B0W660 A0A1Q3F4C3 A0A1Q3F4A8 A0A182GD34 A0A0M8ZXM4 A0A232EX19 K7IRL7 A0A154NXX6 A0A195F0K8 A0A0C9RCV1 A0A1J1HH57 E1ZX65 E9IEA5 A0A0T6B659 A0A195BYY8 A0A158P3K0 A0A1I8M785 A0A1B6CA99 A0A088ANI7 A0A067R7P6 A0A0L7R128 A0A1B6G7E8 A0A2A3EHW7 U5EIA7 A0A2J7R8A3 A0A0J7L9Q7 E2BLZ3 A0A182PNV1 A0A151IH85 A0A182YDY3 A0A151IV51 A0A1B6IYN3 A0A1A9W8R4 A0A1B0DQV1 A0A1A9USZ5 A0A182FLY6 A0A182QS09 A0A1B0FHM2 A0A182NP70 A0A2P8Y1P6 A0A182G371 A0A1A9Z6C1 A0A1L8DWV2 A0A151XDL8 A0A1A9XNK6 F4WAT3 A0A182RWX9 A0A084VJI3 A0A0K8TTP7 A0A1I8NY79 A0A1B0BNV4 W5JCN2 A0A2M4AM18 A0A182W7X2 A0A182LYH5 A0A2M4BRB5 A0A182J789 A0A1B0CFG7 A0A1W4X8C9 A0A182SLD7 A0A1W4X747 A0A0K8W8M5 A0A0L0BZ32 W8CCV9 D2A4G3 A0A2M4BS91 A0A2M4BSI3 A0A2R7VTM9 A0A3B5AIY4 A0A1W5ACE9 A0A1W5AKN7 A0A0A1WTT9 A0A1S3RAU2 A0A0S7LWP7 A0A210PG82 A0A3B3Y372 A0A3Q0JGC9 A0A3B3W2H1 A0A087X2N6 G3Q0C1 A0A3P8RL57 A0A2D0RMY4 A0A315VV76 A0A1Y1NK86 G3Q0C4 A0A3Q2X6R5 A0A3B4TU02 A0A147ARZ8 A0A3Q1IXY4 A0A3S2N614
S4PKV9 Q16TZ7 B0W660 A0A1Q3F4C3 A0A1Q3F4A8 A0A182GD34 A0A0M8ZXM4 A0A232EX19 K7IRL7 A0A154NXX6 A0A195F0K8 A0A0C9RCV1 A0A1J1HH57 E1ZX65 E9IEA5 A0A0T6B659 A0A195BYY8 A0A158P3K0 A0A1I8M785 A0A1B6CA99 A0A088ANI7 A0A067R7P6 A0A0L7R128 A0A1B6G7E8 A0A2A3EHW7 U5EIA7 A0A2J7R8A3 A0A0J7L9Q7 E2BLZ3 A0A182PNV1 A0A151IH85 A0A182YDY3 A0A151IV51 A0A1B6IYN3 A0A1A9W8R4 A0A1B0DQV1 A0A1A9USZ5 A0A182FLY6 A0A182QS09 A0A1B0FHM2 A0A182NP70 A0A2P8Y1P6 A0A182G371 A0A1A9Z6C1 A0A1L8DWV2 A0A151XDL8 A0A1A9XNK6 F4WAT3 A0A182RWX9 A0A084VJI3 A0A0K8TTP7 A0A1I8NY79 A0A1B0BNV4 W5JCN2 A0A2M4AM18 A0A182W7X2 A0A182LYH5 A0A2M4BRB5 A0A182J789 A0A1B0CFG7 A0A1W4X8C9 A0A182SLD7 A0A1W4X747 A0A0K8W8M5 A0A0L0BZ32 W8CCV9 D2A4G3 A0A2M4BS91 A0A2M4BSI3 A0A2R7VTM9 A0A3B5AIY4 A0A1W5ACE9 A0A1W5AKN7 A0A0A1WTT9 A0A1S3RAU2 A0A0S7LWP7 A0A210PG82 A0A3B3Y372 A0A3Q0JGC9 A0A3B3W2H1 A0A087X2N6 G3Q0C1 A0A3P8RL57 A0A2D0RMY4 A0A315VV76 A0A1Y1NK86 G3Q0C4 A0A3Q2X6R5 A0A3B4TU02 A0A147ARZ8 A0A3Q1IXY4 A0A3S2N614
PDB
5HCN
E-value=7.64655e-80,
Score=756
Ontologies
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
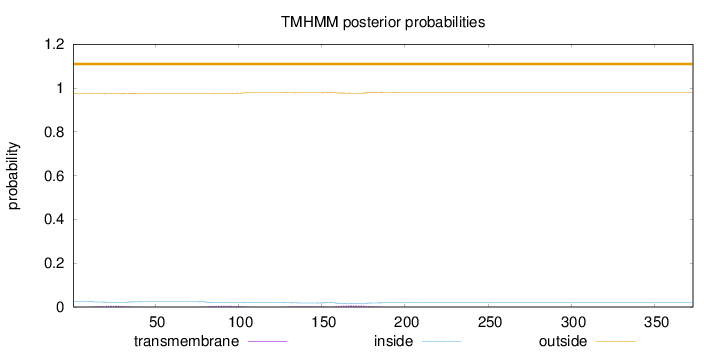
Length:
373
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.41803
Exp number, first 60 AAs:
0.09138
Total prob of N-in:
0.02474
outside
1 - 373
Population Genetic Test Statistics
Pi
2.71193
Theta
18.998912
Tajima's D
-1.605959
CLR
1.326856
CSRT
0.0435978201089946
Interpretation
Uncertain
