Gene
KWMTBOMO13399
Pre Gene Modal
BGIBMGA010907
Annotation
PREDICTED:_FAD_synthase-like_isoform_X1_[Papilio_machaon]
Full name
tRNA (guanine(37)-N1)-methyltransferase
+ More
FAD synthase
FAD synthase
Alternative Name
M1G-methyltransferase
tRNA [GM37] methyltransferase
tRNA methyltransferase 5 homolog
FAD pyrophosphorylase
FMN adenylyltransferase
Flavin adenine dinucleotide synthase
tRNA [GM37] methyltransferase
tRNA methyltransferase 5 homolog
FAD pyrophosphorylase
FMN adenylyltransferase
Flavin adenine dinucleotide synthase
Location in the cell
Cytoplasmic Reliability : 1.414 Extracellular Reliability : 1.165 Nuclear Reliability : 1.314
Sequence
CDS
ATGCAACCGACGTGCGGAGGCATGGACGAGCTACCTGACGTTACTGATGTCCTGAAGGAAGCCGAGCAGGTAATTAGGCAATGCTTCCAAACGTACGCATTGGACGAGGTATTCTTATGTTTCAACGGCGGAAAAGACTGCACAGTATTATTAGACATCACAATAAACGTACTTAAAGATATTTACAAGAGCTGCGACATAGGGAAGAACCTTAAAGTCGTCTACATAAGGACAAAGGGACCGTTTGTCGAAATCGAGAAATTCGTTCAAGAGATTAAAATGTATTATGGTCTGACGTTAAAAGTAACCGAAGGCGAAATGAAGGAGACGTTACAGAGGCTATTAGAGAAGGACGGAATATTGAAGGCCGGCTTGATGGGAACGAGACGGAGCGATCCTTACAGCGAGAATTTGCAATTTGTTCAGAAAACGGATGCGAATTGGCCTCAGATAATGAGAATATCTCCGCTATTGAACTGGTTTTATCACCACATATGGAGCTACATATTGCAACGACAAGTGCCGTACTGCAGTCTCTACGATAAAGGGTACACGTCGATAGGAAGCACAACGAACACGTGGCCGAATCCAGCCTTAGCCCACAAGGACTGCTTCGGTTGCGTCACCTACCACCCCGCCTGGAGATTATCCGACGCGTCCTTAGAGAGAGCCGGTCGAGGAAGTTCAGTGAAGGCACCCGTCAATGGAAACGGTACATATCACAATCATAGTTTGACGTCAACCATCAATCCTTGTATACGCAACGGTGATGTTCTTTGA
Protein
MQPTCGGMDELPDVTDVLKEAEQVIRQCFQTYALDEVFLCFNGGKDCTVLLDITINVLKDIYKSCDIGKNLKVVYIRTKGPFVEIEKFVQEIKMYYGLTLKVTEGEMKETLQRLLEKDGILKAGLMGTRRSDPYSENLQFVQKTDANWPQIMRISPLLNWFYHHIWSYILQRQVPYCSLYDKGYTSIGSTTNTWPNPALAHKDCFGCVTYHPAWRLSDASLERAGRGSSVKAPVNGNGTYHNHSLTSTINPCIRNGDVL
Summary
Description
Specifically methylates the N1 position of guanosine-37 in various cytoplasmic and mitochondrial tRNAs. Methylation is not dependent on the nature of the nucleoside 5' of the target nucleoside. This is the first step in the biosynthesis of wybutosine (yW), a modified base adjacent to the anticodon of tRNAs and required for accurate decoding.
Catalyzes the adenylation of flavin mononucleotide (FMN) to form flavin adenine dinucleotide (FAD) coenzyme.
Catalyzes the adenylation of flavin mononucleotide (FMN) to form flavin adenine dinucleotide (FAD) coenzyme.
Catalytic Activity
guanosine(37) in tRNA + S-adenosyl-L-methionine = H(+) + N(1)-methylguanosine(37) in tRNA + S-adenosyl-L-homocysteine
ATP + FMN + H(+) = diphosphate + FAD
ATP + FMN + H(+) = diphosphate + FAD
Cofactor
Mg(2+)
Subunit
Monomer.
Similarity
Belongs to the TRM5 / TYW2 family.
Belongs to the class I-like SAM-binding methyltransferase superfamily. TRM5/TYW2 family.
Belongs to the AAA ATPase family.
In the C-terminal section; belongs to the PAPS reductase family. FAD1 subfamily.
In the N-terminal section; belongs to the MoaB/Mog family.
Belongs to the class I-like SAM-binding methyltransferase superfamily. TRM5/TYW2 family.
Belongs to the AAA ATPase family.
In the C-terminal section; belongs to the PAPS reductase family. FAD1 subfamily.
In the N-terminal section; belongs to the MoaB/Mog family.
Uniprot
A0A2H1V1S0
A0A194RJ13
S4PD19
A0A212ES28
A0A182MSM2
Q16R18
+ More
A0A182RT01 A0A182JPS7 A0A023EJW0 A0A067RJ45 A0A182UUE1 A0A182HS54 A0A182PTK6 A0A182VZR8 A0A182XAA9 A0A182PUF7 A0A182TEY1 A0A182QMH8 A0A2M4C0J5 A0A2M4ASX7 A0A0T6B8S1 Q7PR81 A0A182LE17 A0A1S3JLB7 W5JFH8 A0A1S3JK05 A0A182FQW9 A0A182Y3X9 A0A1Q3FX95 A0A0P4W0W7 A0A182NQI2 R7TAB2 A0A336KMK0 K1R7T5 A0A084VHR3 A0A1Y1K9C7 S4RHL1 A0A224XBA9 A0A336LH97 F5HKN2 A0A0P4W1D7 A0A1J1IF22 T1GTI8 A0A3R7PN01 B4IEL7 B4Q4T8 V4BL84 A0A0P4W8U6 A0A087UBU6 U4U573 Q8MZ99 Q9VJY1 T1HF00 A0A1W4VKX7 B3MS20 A0A2C9LXT6 E0VLV1 W8CBV0 A0A091I5W9 A0A0P9AA87 A0A3Q3IK62 A0A2C9LXT3 A0A1W4V7U0 B4IGK2 B4P369 A0A210QUG9 A0A0M4EXK7 A0A3M6U4U2 B4M8I2 A0A2Z6SND3 A0A2G8JXV7 A0A2B4RPN7 A0A3Q2QZ39 A0A0P6AJA4 A0A0M8ZRA3 A0A034WDF5 A0A146WT70 A0A2H6N099 A0A146WSC9 U3IP73 A0A372Q6K6 A0A2G8K5S1 A0A0R1E9W5 A0A2P4P0M3 A0A015JDC8 A0A397I2T9 A0A0A1X3K3 A0A146WSL6 A0A3Q0RSU9 A0A0B6Z746 A0A1B6G5V1 B3NAW5 A0A0N8B0I2 V5G434 B4Q1Y5 N6TD15 A0A0K8UPH6 A0A0K2U4F6
A0A182RT01 A0A182JPS7 A0A023EJW0 A0A067RJ45 A0A182UUE1 A0A182HS54 A0A182PTK6 A0A182VZR8 A0A182XAA9 A0A182PUF7 A0A182TEY1 A0A182QMH8 A0A2M4C0J5 A0A2M4ASX7 A0A0T6B8S1 Q7PR81 A0A182LE17 A0A1S3JLB7 W5JFH8 A0A1S3JK05 A0A182FQW9 A0A182Y3X9 A0A1Q3FX95 A0A0P4W0W7 A0A182NQI2 R7TAB2 A0A336KMK0 K1R7T5 A0A084VHR3 A0A1Y1K9C7 S4RHL1 A0A224XBA9 A0A336LH97 F5HKN2 A0A0P4W1D7 A0A1J1IF22 T1GTI8 A0A3R7PN01 B4IEL7 B4Q4T8 V4BL84 A0A0P4W8U6 A0A087UBU6 U4U573 Q8MZ99 Q9VJY1 T1HF00 A0A1W4VKX7 B3MS20 A0A2C9LXT6 E0VLV1 W8CBV0 A0A091I5W9 A0A0P9AA87 A0A3Q3IK62 A0A2C9LXT3 A0A1W4V7U0 B4IGK2 B4P369 A0A210QUG9 A0A0M4EXK7 A0A3M6U4U2 B4M8I2 A0A2Z6SND3 A0A2G8JXV7 A0A2B4RPN7 A0A3Q2QZ39 A0A0P6AJA4 A0A0M8ZRA3 A0A034WDF5 A0A146WT70 A0A2H6N099 A0A146WSC9 U3IP73 A0A372Q6K6 A0A2G8K5S1 A0A0R1E9W5 A0A2P4P0M3 A0A015JDC8 A0A397I2T9 A0A0A1X3K3 A0A146WSL6 A0A3Q0RSU9 A0A0B6Z746 A0A1B6G5V1 B3NAW5 A0A0N8B0I2 V5G434 B4Q1Y5 N6TD15 A0A0K8UPH6 A0A0K2U4F6
EC Number
2.1.1.228
2.7.7.2
2.7.7.2
Pubmed
26354079
23622113
22118469
17510324
24945155
24845553
+ More
12364791 14747013 17210077 20966253 20920257 23761445 25244985 27129103 23254933 22992520 24438588 28004739 17994087 22936249 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 15562597 20566863 24495485 17550304 28812685 30382153 29023486 25348373 24277808 29355972 25830018 24407650
12364791 14747013 17210077 20966253 20920257 23761445 25244985 27129103 23254933 22992520 24438588 28004739 17994087 22936249 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 15562597 20566863 24495485 17550304 28812685 30382153 29023486 25348373 24277808 29355972 25830018 24407650
EMBL
ODYU01000073
SOQ34214.1
KQ460323
KPJ15931.1
GAIX01003831
JAA88729.1
+ More
AGBW02012888 OWR44285.1 AXCM01000621 CH477726 EAT36856.1 GAPW01004226 JAC09372.1 KK852654 KDR19338.1 APCN01000243 AXCN02001534 GGFJ01009367 MBW58508.1 GGFK01010397 MBW43718.1 LJIG01009117 KRT83722.1 AAAB01008859 EAA07691.5 ADMH02001643 ETN61635.1 GFDL01002838 JAV32207.1 GDKW01001249 JAI55346.1 AMQN01003072 KB310836 ELT90649.1 UFQS01000644 UFQT01000644 SSX05689.1 SSX26048.1 JH816527 EKC37260.1 ATLV01013213 KE524845 KFB37507.1 GEZM01088633 JAV58059.1 GFTR01006720 JAW09706.1 UFQS01004045 UFQT01004045 SSX16127.1 SSX35454.1 EGK96784.1 GDRN01080441 JAI62219.1 CVRI01000047 CRK97598.1 CAQQ02030687 QCYY01002259 ROT71711.1 CH480831 EDW46044.1 CM000361 CM002910 EDX04926.1 KMY90101.1 KB202619 ESO89349.1 GDRN01080440 JAI62220.1 KK119125 KFM74835.1 KB631957 ERL87473.1 AY113289 AAM29294.1 AE014134 BT150141 AAF53304.1 AGO63484.1 ACPB03013538 CH902622 EDV34575.1 KPU75203.1 DS235281 EEB14357.1 GAMC01004492 GAMC01004491 JAC02065.1 KL218155 KFP02790.1 KPU75202.1 CH480836 EDW48952.1 CM000157 EDW88311.1 NEDP02001812 OWF52384.1 CP012528 ALC48505.1 RCHS01002246 RMX48606.1 CH940654 EDW57508.1 BEXD01004265 GBC09017.1 MRZV01001112 PIK40525.1 LSMT01000423 PFX18225.1 GDIP01041947 JAM61768.1 KQ435955 KOX68033.1 GAKP01007144 GAKP01007143 JAC51808.1 GCES01054158 JAR32165.1 IACI01027327 LAA21515.1 GCES01054159 JAR32164.1 QKKE01000956 RGB23641.1 MRZV01000861 PIK43299.1 CM000162 KRK06186.1 KRK06187.1 KRK06188.1 AUPC02000486 POG58932.1 JEMT01017717 EXX67487.1 PQFF01000259 RHZ69482.1 GBXI01017523 GBXI01008383 JAC96768.1 JAD05909.1 GCES01054157 JAR32166.1 HACG01016896 CEK63761.1 GECZ01011959 JAS57810.1 CH954177 EDV58679.1 GDIQ01224803 JAK26922.1 BAUL01000283 GAD99193.1 EDX01506.1 APGK01043140 KB741011 ENN75578.1 GDHF01028110 GDHF01026607 GDHF01023727 GDHF01023142 GDHF01009567 JAI24204.1 JAI25707.1 JAI28587.1 JAI29172.1 JAI42747.1 HACA01015450 HACA01015451 CDW32812.1
AGBW02012888 OWR44285.1 AXCM01000621 CH477726 EAT36856.1 GAPW01004226 JAC09372.1 KK852654 KDR19338.1 APCN01000243 AXCN02001534 GGFJ01009367 MBW58508.1 GGFK01010397 MBW43718.1 LJIG01009117 KRT83722.1 AAAB01008859 EAA07691.5 ADMH02001643 ETN61635.1 GFDL01002838 JAV32207.1 GDKW01001249 JAI55346.1 AMQN01003072 KB310836 ELT90649.1 UFQS01000644 UFQT01000644 SSX05689.1 SSX26048.1 JH816527 EKC37260.1 ATLV01013213 KE524845 KFB37507.1 GEZM01088633 JAV58059.1 GFTR01006720 JAW09706.1 UFQS01004045 UFQT01004045 SSX16127.1 SSX35454.1 EGK96784.1 GDRN01080441 JAI62219.1 CVRI01000047 CRK97598.1 CAQQ02030687 QCYY01002259 ROT71711.1 CH480831 EDW46044.1 CM000361 CM002910 EDX04926.1 KMY90101.1 KB202619 ESO89349.1 GDRN01080440 JAI62220.1 KK119125 KFM74835.1 KB631957 ERL87473.1 AY113289 AAM29294.1 AE014134 BT150141 AAF53304.1 AGO63484.1 ACPB03013538 CH902622 EDV34575.1 KPU75203.1 DS235281 EEB14357.1 GAMC01004492 GAMC01004491 JAC02065.1 KL218155 KFP02790.1 KPU75202.1 CH480836 EDW48952.1 CM000157 EDW88311.1 NEDP02001812 OWF52384.1 CP012528 ALC48505.1 RCHS01002246 RMX48606.1 CH940654 EDW57508.1 BEXD01004265 GBC09017.1 MRZV01001112 PIK40525.1 LSMT01000423 PFX18225.1 GDIP01041947 JAM61768.1 KQ435955 KOX68033.1 GAKP01007144 GAKP01007143 JAC51808.1 GCES01054158 JAR32165.1 IACI01027327 LAA21515.1 GCES01054159 JAR32164.1 QKKE01000956 RGB23641.1 MRZV01000861 PIK43299.1 CM000162 KRK06186.1 KRK06187.1 KRK06188.1 AUPC02000486 POG58932.1 JEMT01017717 EXX67487.1 PQFF01000259 RHZ69482.1 GBXI01017523 GBXI01008383 JAC96768.1 JAD05909.1 GCES01054157 JAR32166.1 HACG01016896 CEK63761.1 GECZ01011959 JAS57810.1 CH954177 EDV58679.1 GDIQ01224803 JAK26922.1 BAUL01000283 GAD99193.1 EDX01506.1 APGK01043140 KB741011 ENN75578.1 GDHF01028110 GDHF01026607 GDHF01023727 GDHF01023142 GDHF01009567 JAI24204.1 JAI25707.1 JAI28587.1 JAI29172.1 JAI42747.1 HACA01015450 HACA01015451 CDW32812.1
Proteomes
UP000053240
UP000007151
UP000075883
UP000008820
UP000075900
UP000075881
+ More
UP000027135 UP000075903 UP000075840 UP000075885 UP000075920 UP000076407 UP000075902 UP000075886 UP000007062 UP000075882 UP000085678 UP000000673 UP000069272 UP000076408 UP000075884 UP000014760 UP000005408 UP000030765 UP000245300 UP000183832 UP000015102 UP000283509 UP000001292 UP000000304 UP000030746 UP000054359 UP000030742 UP000000803 UP000015103 UP000192221 UP000007801 UP000076420 UP000009046 UP000054308 UP000261600 UP000002282 UP000242188 UP000092553 UP000275408 UP000008792 UP000247702 UP000230750 UP000225706 UP000265000 UP000053105 UP000263633 UP000018888 UP000022910 UP000266861 UP000261340 UP000008711 UP000018001 UP000019118
UP000027135 UP000075903 UP000075840 UP000075885 UP000075920 UP000076407 UP000075902 UP000075886 UP000007062 UP000075882 UP000085678 UP000000673 UP000069272 UP000076408 UP000075884 UP000014760 UP000005408 UP000030765 UP000245300 UP000183832 UP000015102 UP000283509 UP000001292 UP000000304 UP000030746 UP000054359 UP000030742 UP000000803 UP000015103 UP000192221 UP000007801 UP000076420 UP000009046 UP000054308 UP000261600 UP000002282 UP000242188 UP000092553 UP000275408 UP000008792 UP000247702 UP000230750 UP000225706 UP000265000 UP000053105 UP000263633 UP000018888 UP000022910 UP000266861 UP000261340 UP000008711 UP000018001 UP000019118
Interpro
IPR030382
MeTrfase_TRM5/TYW2
+ More
IPR025792 tRNA_Gua_MeTrfase_euk
IPR029063 SAM-dependent_MTases
IPR002500 PAPS_reduct
IPR014729 Rossmann-like_a/b/a_fold
IPR001453 MoaB/Mog_dom
IPR036425 MoaB/Mog-like_dom_sf
IPR003960 ATPase_AAA_CS
IPR027417 P-loop_NTPase
IPR003959 ATPase_AAA_core
IPR005937 26S_Psome_P45-like
IPR003593 AAA+_ATPase
IPR041569 AAA_lid_3
IPR035245 PSMC2
IPR012183 FAD_synth_MoaB/Mog-bd
IPR025792 tRNA_Gua_MeTrfase_euk
IPR029063 SAM-dependent_MTases
IPR002500 PAPS_reduct
IPR014729 Rossmann-like_a/b/a_fold
IPR001453 MoaB/Mog_dom
IPR036425 MoaB/Mog-like_dom_sf
IPR003960 ATPase_AAA_CS
IPR027417 P-loop_NTPase
IPR003959 ATPase_AAA_core
IPR005937 26S_Psome_P45-like
IPR003593 AAA+_ATPase
IPR041569 AAA_lid_3
IPR035245 PSMC2
IPR012183 FAD_synth_MoaB/Mog-bd
Gene 3D
ProteinModelPortal
A0A2H1V1S0
A0A194RJ13
S4PD19
A0A212ES28
A0A182MSM2
Q16R18
+ More
A0A182RT01 A0A182JPS7 A0A023EJW0 A0A067RJ45 A0A182UUE1 A0A182HS54 A0A182PTK6 A0A182VZR8 A0A182XAA9 A0A182PUF7 A0A182TEY1 A0A182QMH8 A0A2M4C0J5 A0A2M4ASX7 A0A0T6B8S1 Q7PR81 A0A182LE17 A0A1S3JLB7 W5JFH8 A0A1S3JK05 A0A182FQW9 A0A182Y3X9 A0A1Q3FX95 A0A0P4W0W7 A0A182NQI2 R7TAB2 A0A336KMK0 K1R7T5 A0A084VHR3 A0A1Y1K9C7 S4RHL1 A0A224XBA9 A0A336LH97 F5HKN2 A0A0P4W1D7 A0A1J1IF22 T1GTI8 A0A3R7PN01 B4IEL7 B4Q4T8 V4BL84 A0A0P4W8U6 A0A087UBU6 U4U573 Q8MZ99 Q9VJY1 T1HF00 A0A1W4VKX7 B3MS20 A0A2C9LXT6 E0VLV1 W8CBV0 A0A091I5W9 A0A0P9AA87 A0A3Q3IK62 A0A2C9LXT3 A0A1W4V7U0 B4IGK2 B4P369 A0A210QUG9 A0A0M4EXK7 A0A3M6U4U2 B4M8I2 A0A2Z6SND3 A0A2G8JXV7 A0A2B4RPN7 A0A3Q2QZ39 A0A0P6AJA4 A0A0M8ZRA3 A0A034WDF5 A0A146WT70 A0A2H6N099 A0A146WSC9 U3IP73 A0A372Q6K6 A0A2G8K5S1 A0A0R1E9W5 A0A2P4P0M3 A0A015JDC8 A0A397I2T9 A0A0A1X3K3 A0A146WSL6 A0A3Q0RSU9 A0A0B6Z746 A0A1B6G5V1 B3NAW5 A0A0N8B0I2 V5G434 B4Q1Y5 N6TD15 A0A0K8UPH6 A0A0K2U4F6
A0A182RT01 A0A182JPS7 A0A023EJW0 A0A067RJ45 A0A182UUE1 A0A182HS54 A0A182PTK6 A0A182VZR8 A0A182XAA9 A0A182PUF7 A0A182TEY1 A0A182QMH8 A0A2M4C0J5 A0A2M4ASX7 A0A0T6B8S1 Q7PR81 A0A182LE17 A0A1S3JLB7 W5JFH8 A0A1S3JK05 A0A182FQW9 A0A182Y3X9 A0A1Q3FX95 A0A0P4W0W7 A0A182NQI2 R7TAB2 A0A336KMK0 K1R7T5 A0A084VHR3 A0A1Y1K9C7 S4RHL1 A0A224XBA9 A0A336LH97 F5HKN2 A0A0P4W1D7 A0A1J1IF22 T1GTI8 A0A3R7PN01 B4IEL7 B4Q4T8 V4BL84 A0A0P4W8U6 A0A087UBU6 U4U573 Q8MZ99 Q9VJY1 T1HF00 A0A1W4VKX7 B3MS20 A0A2C9LXT6 E0VLV1 W8CBV0 A0A091I5W9 A0A0P9AA87 A0A3Q3IK62 A0A2C9LXT3 A0A1W4V7U0 B4IGK2 B4P369 A0A210QUG9 A0A0M4EXK7 A0A3M6U4U2 B4M8I2 A0A2Z6SND3 A0A2G8JXV7 A0A2B4RPN7 A0A3Q2QZ39 A0A0P6AJA4 A0A0M8ZRA3 A0A034WDF5 A0A146WT70 A0A2H6N099 A0A146WSC9 U3IP73 A0A372Q6K6 A0A2G8K5S1 A0A0R1E9W5 A0A2P4P0M3 A0A015JDC8 A0A397I2T9 A0A0A1X3K3 A0A146WSL6 A0A3Q0RSU9 A0A0B6Z746 A0A1B6G5V1 B3NAW5 A0A0N8B0I2 V5G434 B4Q1Y5 N6TD15 A0A0K8UPH6 A0A0K2U4F6
PDB
3G6K
E-value=4.10112e-25,
Score=282
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Predominantly in the mitochondria and in the nucleus. With evidence from 1 publications.
Mitochondrion matrix Predominantly in the mitochondria and in the nucleus. With evidence from 1 publications.
Nucleus Predominantly in the mitochondria and in the nucleus. With evidence from 1 publications.
Mitochondrion matrix Predominantly in the mitochondria and in the nucleus. With evidence from 1 publications.
Nucleus Predominantly in the mitochondria and in the nucleus. With evidence from 1 publications.
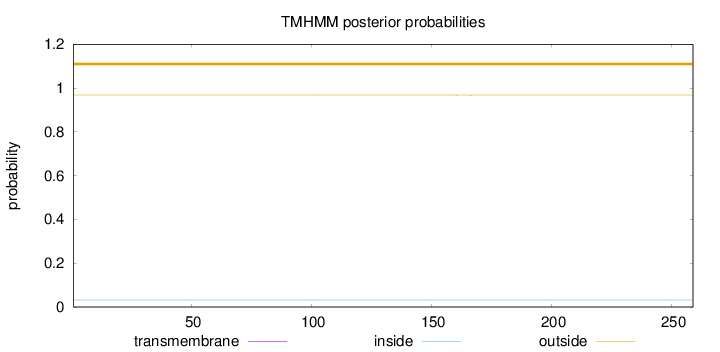
Length:
259
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00987
Exp number, first 60 AAs:
0.00088
Total prob of N-in:
0.03188
outside
1 - 259
Population Genetic Test Statistics
Pi
37.495695
Theta
25.781048
Tajima's D
-1.106918
CLR
1.244701
CSRT
0.11624418779061
Interpretation
Uncertain
