Gene
KWMTBOMO13398
Pre Gene Modal
BGIBMGA010908
Annotation
PREDICTED:_tRNA_(guanine(37)-N1)-methyltransferase_[Amyelois_transitella]
Full name
tRNA (guanine(37)-N1)-methyltransferase
Alternative Name
M1G-methyltransferase
tRNA [GM37] methyltransferase
tRNA methyltransferase 5 homolog
tRNA [GM37] methyltransferase
tRNA methyltransferase 5 homolog
Location in the cell
Nuclear Reliability : 2.887
Sequence
CDS
ATGACTTCCTCTTTGGTTCCCCTCAATGTTCGTGGTATGAAGGTCCTTGACCGCTCAAAATTCTTGCGTAACATCAAAGTGCCAGTGTTGAAGGTCAGTGAACAGACATTGGCGAAAATTACACCACTCTGCAAGTCTTACTTTTTAAAACTGGAACGATTCAAGCCGGTACAGAGCTCGAAGCTGGACGGAGAGGAGTCCAAAAAAAACATTTACTTGAACCCTGATACTATAAACAGTTGGACTGATATTGAAGAAAATGTTCGGAAGGACATGTCCGTCTTCGAGGTCAATGAATCCAACTTTAATACCCAAGAAATACAACTGACATACGATAATTGGAGTTTTGACATCATTTTCAATGCTGTCTTACCTGAAAATGAAGAAAAACTAAGCAGTTTCTCACACATCGGACACATCATTCACTTGAATTTACGAGATCATTTACTTGATTATCGTTTCTTGATTGGTGAAGTCTTGCTCGACAAAATAAGGTCATGTCGGACTGTAGTCAATAAAAGTAACACTATAAATAATACATATCGCAACTTCAGCATGGAACTTTTAGCAGGGGACGAAGATTATATTGTTACTGTCAAAGAAAATCGATGTAATTTCGAATTTGATTTCTCAAAAGTTTACTGGAATCCAAGATTATGTAAAGAGCACGAAAGAATACTAAGTTATTTGAAACAAGGCGATGTCTTATTTGACATCTTCTGTGGAGTTGGACCATTCTCGATACCAGCAGCGAAACGCAAAACCATTGTTTATGCAAACGACCTGAATCCTGAATCATACAAATGGTTGAACCATAATGCAGAGAAAAATAAAATAAACAATCAATACTTTAAATCATACAATTTGGATGGTAAGGAATTCATTTTAACTGTGCTCAAAGATTATTTATTAGATCTATGTAATGAAAAAAGAAAGCTCGATGATAGTTCTCGGATACACATTACCATGAATCTACCAGCATTGGCAGTTGAATTTTTGAAATATTTTAAGGGTTTGGTGCGTAGTGAGAATGTAAATAATTTGAAAAATGAAATTATAGTTTATGTTTATTGTTTTGCGGCTACAAAGGAAATTGCAAAGGAAATGGTCTGTGAAAATATTGCCATTGATATCTCAAAGAATATAATTGAAGTGTTTGATGTACGTAATGTGTCTCCAAAGAAGGAGATGATGCGTGTTACCATTAAGTTAACTAATGATATTTTGTGCATAAATAGTGATGATCATAGTGAACCTCCTTTGAAAAAGTTATGTGTTGAAAGTAAATAG
Protein
MTSSLVPLNVRGMKVLDRSKFLRNIKVPVLKVSEQTLAKITPLCKSYFLKLERFKPVQSSKLDGEESKKNIYLNPDTINSWTDIEENVRKDMSVFEVNESNFNTQEIQLTYDNWSFDIIFNAVLPENEEKLSSFSHIGHIIHLNLRDHLLDYRFLIGEVLLDKIRSCRTVVNKSNTINNTYRNFSMELLAGDEDYIVTVKENRCNFEFDFSKVYWNPRLCKEHERILSYLKQGDVLFDIFCGVGPFSIPAAKRKTIVYANDLNPESYKWLNHNAEKNKINNQYFKSYNLDGKEFILTVLKDYLLDLCNEKRKLDDSSRIHITMNLPALAVEFLKYFKGLVRSENVNNLKNEIIVYVYCFAATKEIAKEMVCENIAIDISKNIIEVFDVRNVSPKKEMMRVTIKLTNDILCINSDDHSEPPLKKLCVESK
Summary
Description
Specifically methylates the N1 position of guanosine-37 in various cytoplasmic and mitochondrial tRNAs. Methylation is not dependent on the nature of the nucleoside 5' of the target nucleoside. This is the first step in the biosynthesis of wybutosine (yW), a modified base adjacent to the anticodon of tRNAs and required for accurate decoding.
Catalytic Activity
guanosine(37) in tRNA + S-adenosyl-L-methionine = H(+) + N(1)-methylguanosine(37) in tRNA + S-adenosyl-L-homocysteine
Subunit
Monomer.
Similarity
Belongs to the TRM5 / TYW2 family.
Belongs to the class I-like SAM-binding methyltransferase superfamily. TRM5/TYW2 family.
Belongs to the class I-like SAM-binding methyltransferase superfamily. TRM5/TYW2 family.
Keywords
Complete proteome
Cytoplasm
Methyltransferase
Mitochondrion
Nucleus
Reference proteome
S-adenosyl-L-methionine
Transferase
tRNA processing
Transit peptide
Feature
chain tRNA (guanine(37)-N1)-methyltransferase
Uniprot
H9JN05
A0A2H1V1S0
A0A194RF52
A0A194Q3T7
A0A212ES03
A0A3S2NRD2
+ More
A0A0L7KT62 A0A067R9N8 A0A1I8PCU9 B4N3S3 A0A2Y9D0S2 B4KXG2 A0A1Y1KV63 A0A0K8UMH9 A0A0M5J048 A0A0K8V2V0 B5DPF1 A0A1C7CZE4 A0A084VW66 B3M7D1 A0A1W4W1D7 B4IX34 B4PGK0 A0A182UFH7 Q7Q5Z3 A0A182GC53 N6SWZ3 Q8IRE4 A0A1I8NEJ3 B4LFT5 B4GRQ9 A0A1A9VJ48 A0A1W4X5K3 B4QP31 A0A182Y2T0 B3NFW6 A0A1B0AEX2 A0A182PDT7 U4TY66 B4HTL3 A0A3B0JU09 A0A164Y9N5 A0A182IV32 A0A182SCD1 A0A336MG57 E9FY52 A0A0P5S8B3 A0A0P6D5Z4 A0A182Q688 A0A182LVP9 A0A0P6E0T2 A0A0P6BQ20 A0A1S4FMY3 A0A336MG40 A0A182JPT4 Q16VC0 A0A1A9XV49 A0A182FGN2 A0A0P5K8R5 A0A0P5FYT4 A0A336M7L9 A0A2J7Q8P6 A0A0P6H6D6 A0A0A1XLG3 A0A1B0FCA8 A0A0A1WZL1 E3WPP8 A0A0L0CA35 A0A0P6EC10 A0A1B0BHD5 W8C5M3 B7T4L2 D7EJV3 A0A0N8DBQ7 A0A0P5UMZ5 A0A1P1DQF0 B0WR25 A0A1B0CKP8 A0A1B6K7K8 A0A182WJY5 A0A2P6L3J5 A0A1B0DRL0 A0A0P5IZ84 R7TC87 E0VLV0 A0A131XBP7 A0A0C9RNW7 A0A0J7KJ60 A0A232F970 A0A195ED18 A0A1Z5LCA8 W5LPT7 L7LZN1 A0A2D0QNI8 A0A3B5A9T5 A0A1W4Y3Y3 A0A3Q1FA53 A0A3L8DCJ5 A0A1S3IL28
A0A0L7KT62 A0A067R9N8 A0A1I8PCU9 B4N3S3 A0A2Y9D0S2 B4KXG2 A0A1Y1KV63 A0A0K8UMH9 A0A0M5J048 A0A0K8V2V0 B5DPF1 A0A1C7CZE4 A0A084VW66 B3M7D1 A0A1W4W1D7 B4IX34 B4PGK0 A0A182UFH7 Q7Q5Z3 A0A182GC53 N6SWZ3 Q8IRE4 A0A1I8NEJ3 B4LFT5 B4GRQ9 A0A1A9VJ48 A0A1W4X5K3 B4QP31 A0A182Y2T0 B3NFW6 A0A1B0AEX2 A0A182PDT7 U4TY66 B4HTL3 A0A3B0JU09 A0A164Y9N5 A0A182IV32 A0A182SCD1 A0A336MG57 E9FY52 A0A0P5S8B3 A0A0P6D5Z4 A0A182Q688 A0A182LVP9 A0A0P6E0T2 A0A0P6BQ20 A0A1S4FMY3 A0A336MG40 A0A182JPT4 Q16VC0 A0A1A9XV49 A0A182FGN2 A0A0P5K8R5 A0A0P5FYT4 A0A336M7L9 A0A2J7Q8P6 A0A0P6H6D6 A0A0A1XLG3 A0A1B0FCA8 A0A0A1WZL1 E3WPP8 A0A0L0CA35 A0A0P6EC10 A0A1B0BHD5 W8C5M3 B7T4L2 D7EJV3 A0A0N8DBQ7 A0A0P5UMZ5 A0A1P1DQF0 B0WR25 A0A1B0CKP8 A0A1B6K7K8 A0A182WJY5 A0A2P6L3J5 A0A1B0DRL0 A0A0P5IZ84 R7TC87 E0VLV0 A0A131XBP7 A0A0C9RNW7 A0A0J7KJ60 A0A232F970 A0A195ED18 A0A1Z5LCA8 W5LPT7 L7LZN1 A0A2D0QNI8 A0A3B5A9T5 A0A1W4Y3Y3 A0A3Q1FA53 A0A3L8DCJ5 A0A1S3IL28
EC Number
2.1.1.228
Pubmed
19121390
26354079
22118469
26227816
24845553
17994087
+ More
28004739 15632085 20966253 24438588 17550304 12364791 26483478 23537049 10731132 12537572 25315136 22936249 25244985 21292972 17510324 25830018 20920257 26108605 24495485 19061532 18362917 19820115 23254933 20566863 28049606 28648823 28528879 25329095 25576852 30249741
28004739 15632085 20966253 24438588 17550304 12364791 26483478 23537049 10731132 12537572 25315136 22936249 25244985 21292972 17510324 25830018 20920257 26108605 24495485 19061532 18362917 19820115 23254933 20566863 28049606 28648823 28528879 25329095 25576852 30249741
EMBL
BABH01041387
ODYU01000073
SOQ34214.1
KQ460323
KPJ15930.1
KQ459586
+ More
KPI98070.1 AGBW02012888 OWR44286.1 RSAL01000124 RVE46611.1 JTDY01006198 KOB66204.1 KK852654 KDR19337.1 CH964095 EDW79278.2 APCN01000054 CH933809 EDW18648.2 GEZM01077469 JAV63296.1 GDHF01024611 JAI27703.1 CP012525 ALC42936.1 GDHF01019095 JAI33219.1 CH379069 ATLV01017495 KE525170 KFB42210.1 CH902618 EDV39829.2 CH916366 EDV96340.1 CM000159 EDW93218.1 AAAB01008960 JXUM01009387 KQ560282 KXJ83316.1 APGK01053536 KB741224 ENN72309.1 AE014296 BT082011 CH940647 EDW69312.2 CH479188 EDW40444.1 CM000363 CM002912 EDX09026.1 KMY97277.1 CH954178 EDV50728.1 KB631749 ERL85757.1 CH480817 EDW50284.1 OUUW01000002 SPP76201.1 LRGB01000930 KZS15027.1 UFQT01001153 SSX29176.1 GL732527 EFX87837.1 GDIQ01099848 JAL51878.1 GDIQ01082291 JAN12446.1 AXCN02000962 AXCM01000220 GDIQ01074102 JAN20635.1 GDIP01011475 JAM92240.1 UFQS01001153 SSX09273.1 SSX29175.1 CH477597 GDIQ01193121 JAK58604.1 GDIQ01253802 JAJ97922.1 UFQS01000664 UFQT01000664 SSX05907.1 SSX26266.1 NEVH01016949 PNF24943.1 GDIQ01036589 JAN58148.1 GBXI01002491 JAD11801.1 CCAG010008817 GBXI01013765 GBXI01010327 JAD00527.1 JAD03965.1 ADMH02002104 ETN59038.1 JRES01000692 KNC29111.1 GDIQ01066263 JAN28474.1 JXJN01014265 GAMC01004774 JAC01782.1 EU931152 ACK57236.1 KQ971866 EFA12884.2 GDIP01049391 JAM54324.1 GDIP01128182 JAL75532.1 DS232049 EDS33123.1 AJWK01016511 AJWK01016512 AJWK01016513 GECU01000266 JAT07441.1 MWRG01002050 PRD33151.1 AJVK01000468 GDIQ01208410 JAK43315.1 AMQN01013948 KB310625 ELT91137.1 DS235281 GEFH01005046 JAP63535.1 GBYB01010135 GBYB01010136 JAG79902.1 JAG79903.1 LBMM01006611 KMQ90463.1 NNAY01000681 OXU27008.1 KQ979074 KYN22739.1 GFJQ02001924 JAW05046.1 GACK01007784 JAA57250.1 QOIP01000010 RLU18051.1
KPI98070.1 AGBW02012888 OWR44286.1 RSAL01000124 RVE46611.1 JTDY01006198 KOB66204.1 KK852654 KDR19337.1 CH964095 EDW79278.2 APCN01000054 CH933809 EDW18648.2 GEZM01077469 JAV63296.1 GDHF01024611 JAI27703.1 CP012525 ALC42936.1 GDHF01019095 JAI33219.1 CH379069 ATLV01017495 KE525170 KFB42210.1 CH902618 EDV39829.2 CH916366 EDV96340.1 CM000159 EDW93218.1 AAAB01008960 JXUM01009387 KQ560282 KXJ83316.1 APGK01053536 KB741224 ENN72309.1 AE014296 BT082011 CH940647 EDW69312.2 CH479188 EDW40444.1 CM000363 CM002912 EDX09026.1 KMY97277.1 CH954178 EDV50728.1 KB631749 ERL85757.1 CH480817 EDW50284.1 OUUW01000002 SPP76201.1 LRGB01000930 KZS15027.1 UFQT01001153 SSX29176.1 GL732527 EFX87837.1 GDIQ01099848 JAL51878.1 GDIQ01082291 JAN12446.1 AXCN02000962 AXCM01000220 GDIQ01074102 JAN20635.1 GDIP01011475 JAM92240.1 UFQS01001153 SSX09273.1 SSX29175.1 CH477597 GDIQ01193121 JAK58604.1 GDIQ01253802 JAJ97922.1 UFQS01000664 UFQT01000664 SSX05907.1 SSX26266.1 NEVH01016949 PNF24943.1 GDIQ01036589 JAN58148.1 GBXI01002491 JAD11801.1 CCAG010008817 GBXI01013765 GBXI01010327 JAD00527.1 JAD03965.1 ADMH02002104 ETN59038.1 JRES01000692 KNC29111.1 GDIQ01066263 JAN28474.1 JXJN01014265 GAMC01004774 JAC01782.1 EU931152 ACK57236.1 KQ971866 EFA12884.2 GDIP01049391 JAM54324.1 GDIP01128182 JAL75532.1 DS232049 EDS33123.1 AJWK01016511 AJWK01016512 AJWK01016513 GECU01000266 JAT07441.1 MWRG01002050 PRD33151.1 AJVK01000468 GDIQ01208410 JAK43315.1 AMQN01013948 KB310625 ELT91137.1 DS235281 GEFH01005046 JAP63535.1 GBYB01010135 GBYB01010136 JAG79902.1 JAG79903.1 LBMM01006611 KMQ90463.1 NNAY01000681 OXU27008.1 KQ979074 KYN22739.1 GFJQ02001924 JAW05046.1 GACK01007784 JAA57250.1 QOIP01000010 RLU18051.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000283053
UP000037510
+ More
UP000027135 UP000095300 UP000007798 UP000075840 UP000009192 UP000092553 UP000001819 UP000075882 UP000030765 UP000007801 UP000192221 UP000001070 UP000002282 UP000075902 UP000007062 UP000069940 UP000249989 UP000019118 UP000000803 UP000095301 UP000008792 UP000008744 UP000078200 UP000192223 UP000000304 UP000076408 UP000008711 UP000092445 UP000075885 UP000030742 UP000001292 UP000268350 UP000076858 UP000075880 UP000075901 UP000000305 UP000075886 UP000075883 UP000075881 UP000008820 UP000092443 UP000069272 UP000235965 UP000092444 UP000000673 UP000037069 UP000092460 UP000007266 UP000002320 UP000092461 UP000075920 UP000092462 UP000014760 UP000009046 UP000036403 UP000215335 UP000078492 UP000018467 UP000221080 UP000261400 UP000192224 UP000257200 UP000279307 UP000085678
UP000027135 UP000095300 UP000007798 UP000075840 UP000009192 UP000092553 UP000001819 UP000075882 UP000030765 UP000007801 UP000192221 UP000001070 UP000002282 UP000075902 UP000007062 UP000069940 UP000249989 UP000019118 UP000000803 UP000095301 UP000008792 UP000008744 UP000078200 UP000192223 UP000000304 UP000076408 UP000008711 UP000092445 UP000075885 UP000030742 UP000001292 UP000268350 UP000076858 UP000075880 UP000075901 UP000000305 UP000075886 UP000075883 UP000075881 UP000008820 UP000092443 UP000069272 UP000235965 UP000092444 UP000000673 UP000037069 UP000092460 UP000007266 UP000002320 UP000092461 UP000075920 UP000092462 UP000014760 UP000009046 UP000036403 UP000215335 UP000078492 UP000018467 UP000221080 UP000261400 UP000192224 UP000257200 UP000279307 UP000085678
Interpro
IPR030382
MeTrfase_TRM5/TYW2
+ More
IPR025792 tRNA_Gua_MeTrfase_euk
IPR029063 SAM-dependent_MTases
IPR002500 PAPS_reduct
IPR014729 Rossmann-like_a/b/a_fold
IPR031389 DUF4665
IPR015919 Cadherin-like_sf
IPR020894 Cadherin_CS
IPR036877 SUI1_dom_sf
IPR005873 DENR_eukaryotes
IPR001950 SUI1
IPR002126 Cadherin-like_dom
IPR025792 tRNA_Gua_MeTrfase_euk
IPR029063 SAM-dependent_MTases
IPR002500 PAPS_reduct
IPR014729 Rossmann-like_a/b/a_fold
IPR031389 DUF4665
IPR015919 Cadherin-like_sf
IPR020894 Cadherin_CS
IPR036877 SUI1_dom_sf
IPR005873 DENR_eukaryotes
IPR001950 SUI1
IPR002126 Cadherin-like_dom
Gene 3D
ProteinModelPortal
H9JN05
A0A2H1V1S0
A0A194RF52
A0A194Q3T7
A0A212ES03
A0A3S2NRD2
+ More
A0A0L7KT62 A0A067R9N8 A0A1I8PCU9 B4N3S3 A0A2Y9D0S2 B4KXG2 A0A1Y1KV63 A0A0K8UMH9 A0A0M5J048 A0A0K8V2V0 B5DPF1 A0A1C7CZE4 A0A084VW66 B3M7D1 A0A1W4W1D7 B4IX34 B4PGK0 A0A182UFH7 Q7Q5Z3 A0A182GC53 N6SWZ3 Q8IRE4 A0A1I8NEJ3 B4LFT5 B4GRQ9 A0A1A9VJ48 A0A1W4X5K3 B4QP31 A0A182Y2T0 B3NFW6 A0A1B0AEX2 A0A182PDT7 U4TY66 B4HTL3 A0A3B0JU09 A0A164Y9N5 A0A182IV32 A0A182SCD1 A0A336MG57 E9FY52 A0A0P5S8B3 A0A0P6D5Z4 A0A182Q688 A0A182LVP9 A0A0P6E0T2 A0A0P6BQ20 A0A1S4FMY3 A0A336MG40 A0A182JPT4 Q16VC0 A0A1A9XV49 A0A182FGN2 A0A0P5K8R5 A0A0P5FYT4 A0A336M7L9 A0A2J7Q8P6 A0A0P6H6D6 A0A0A1XLG3 A0A1B0FCA8 A0A0A1WZL1 E3WPP8 A0A0L0CA35 A0A0P6EC10 A0A1B0BHD5 W8C5M3 B7T4L2 D7EJV3 A0A0N8DBQ7 A0A0P5UMZ5 A0A1P1DQF0 B0WR25 A0A1B0CKP8 A0A1B6K7K8 A0A182WJY5 A0A2P6L3J5 A0A1B0DRL0 A0A0P5IZ84 R7TC87 E0VLV0 A0A131XBP7 A0A0C9RNW7 A0A0J7KJ60 A0A232F970 A0A195ED18 A0A1Z5LCA8 W5LPT7 L7LZN1 A0A2D0QNI8 A0A3B5A9T5 A0A1W4Y3Y3 A0A3Q1FA53 A0A3L8DCJ5 A0A1S3IL28
A0A0L7KT62 A0A067R9N8 A0A1I8PCU9 B4N3S3 A0A2Y9D0S2 B4KXG2 A0A1Y1KV63 A0A0K8UMH9 A0A0M5J048 A0A0K8V2V0 B5DPF1 A0A1C7CZE4 A0A084VW66 B3M7D1 A0A1W4W1D7 B4IX34 B4PGK0 A0A182UFH7 Q7Q5Z3 A0A182GC53 N6SWZ3 Q8IRE4 A0A1I8NEJ3 B4LFT5 B4GRQ9 A0A1A9VJ48 A0A1W4X5K3 B4QP31 A0A182Y2T0 B3NFW6 A0A1B0AEX2 A0A182PDT7 U4TY66 B4HTL3 A0A3B0JU09 A0A164Y9N5 A0A182IV32 A0A182SCD1 A0A336MG57 E9FY52 A0A0P5S8B3 A0A0P6D5Z4 A0A182Q688 A0A182LVP9 A0A0P6E0T2 A0A0P6BQ20 A0A1S4FMY3 A0A336MG40 A0A182JPT4 Q16VC0 A0A1A9XV49 A0A182FGN2 A0A0P5K8R5 A0A0P5FYT4 A0A336M7L9 A0A2J7Q8P6 A0A0P6H6D6 A0A0A1XLG3 A0A1B0FCA8 A0A0A1WZL1 E3WPP8 A0A0L0CA35 A0A0P6EC10 A0A1B0BHD5 W8C5M3 B7T4L2 D7EJV3 A0A0N8DBQ7 A0A0P5UMZ5 A0A1P1DQF0 B0WR25 A0A1B0CKP8 A0A1B6K7K8 A0A182WJY5 A0A2P6L3J5 A0A1B0DRL0 A0A0P5IZ84 R7TC87 E0VLV0 A0A131XBP7 A0A0C9RNW7 A0A0J7KJ60 A0A232F970 A0A195ED18 A0A1Z5LCA8 W5LPT7 L7LZN1 A0A2D0QNI8 A0A3B5A9T5 A0A1W4Y3Y3 A0A3Q1FA53 A0A3L8DCJ5 A0A1S3IL28
PDB
5YAC
E-value=7.58252e-20,
Score=239
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Predominantly in the mitochondria and in the nucleus. With evidence from 1 publications.
Mitochondrion matrix Predominantly in the mitochondria and in the nucleus. With evidence from 1 publications.
Nucleus Predominantly in the mitochondria and in the nucleus. With evidence from 1 publications.
Mitochondrion matrix Predominantly in the mitochondria and in the nucleus. With evidence from 1 publications.
Nucleus Predominantly in the mitochondria and in the nucleus. With evidence from 1 publications.
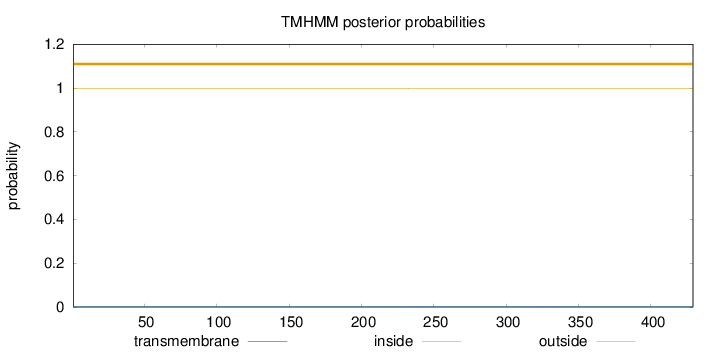
Length:
429
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01171
Exp number, first 60 AAs:
0.00059
Total prob of N-in:
0.00170
outside
1 - 429
Population Genetic Test Statistics
Pi
32.664507
Theta
23.48695
Tajima's D
1.195102
CLR
0
CSRT
0.718414079296035
Interpretation
Uncertain
