Gene
KWMTBOMO13391
Pre Gene Modal
BGIBMGA010938
Annotation
PREDICTED:_proton-coupled_folate_transporter-like_[Papilio_machaon]
Full name
Angiotensin-converting enzyme
Location in the cell
PlasmaMembrane Reliability : 4.212
Sequence
CDS
ATGGGAGCTTTCGTATACGCTTTCGCCAGGAACAACTTGGAGATATATATCGCTCCGTTGATGGAAATTTTAAATGGAACATCATTCATAGCGATGCGATCGATCGCAACGAAACTGGTGAGCGGGGAGGAATTTGGCAAGGTGAATTCTTTGTTCGGTCTGGCGGAGGCGATGATGCCGCTGGTTTACGGACCTCTCTACTCCCGCGTCTACATGGCGACGCTCAACGTGCTGCCAGGCGCTGTCTTCCTGCTCGGAGCCAGTTTGACTTTACCTGCTATTTTCATATTTGGGTGGTTGTATTTTGAACATAAAAAAGACGCACAGAGGAAAGTAAACACAACATGTGATTCGGAAAAATAA
Protein
MGAFVYAFARNNLEIYIAPLMEILNGTSFIAMRSIATKLVSGEEFGKVNSLFGLAEAMMPLVYGPLYSRVYMATLNVLPGAVFLLGASLTLPAIFIFGWLYFEHKKDAQRKVNTTCDSEK
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Belongs to the peptidase M2 family.
Belongs to the peptidase M2 family.
Uniprot
A0A194RE89
A0A194PXP8
A0A3S2PBF1
A0A212FAJ8
A0A212FAK5
A0A194REM1
+ More
A0A2H1X2C4 A0A2P8YGX5 A0A3S2LH66 A0A1B6GHB7 A0A2A4JD86 A0A1B6HTG8 A0A2A4IY18 A0A194PXM6 A0A1B6CY04 D2A535 A0A2S2NH85 A0A1B6LYH9 A0A0J7KQN9 A0A1J1HPT6 A0A0K8SJM1 A0A154PAY0 A0A1W4WW74 A0A1Y1KF48 A0A0A9WCX6 A0A2H1WT49 A0A0T6BFY6 A0A2S2PZG0 A0A146LMI6 A0A023F3V3 A0A069DVE7 A0A194QS69 A0A1E1W1C8 A0A2M4AAV7 A0A2M4CTY3 A0A2M4CUE7 A0A2M4BIX1 A0A2M4CSN5 A0A2M4BIV5 A0A2M4CUE3 A0A212FAL1 J9KB87 A0A2H8TM94 Q16W79 A0A2M3Z330 A0A2M3Z363 U4URR0 A0A1Q3FL61 A0A1Q3FKN7 B0WAI1 E2BSZ7 A0A1Q3FLC7 A0A1Q3FLB8 A0A182QCE8 A0A026WMI6 A0A2A4IXD4 A0A182J2E5 A0A0K8VHS8 A0A2R7W706 A0A1L8DE03 A0A182MSS2 A0A182VHT3 A0A2C9H8S5 A0A1Y1KH29 Q5TSW5 A0A1L8DDP9 F4WT02 A0A151ITA2 A0A158NYI1 A0A195AZB4 A0A151WHP4 E2A659 A0A195CVZ7 A0A195F6C4 A0A3S2LGU6 A0A0L7RGJ1 A0A182H1C2 A0A2S2NPL3 A0A084WT83 A0A182N9U1 E9IUT7 A0A194QAI0 H9JAJ5 A0A0M8ZQJ4 A0A212EJ99 A0A182K2U1 A0A2J7PUC7 A0A194RIY8 A0A2H8THB2 A0A182RDY9 A0A182PQS5 A0A182W3L5 A0A0K8WKB4 A0A2W1BZD6 A0A088ANF5 A0A2A3EDD3 D6WRR1 A0A034W601 A0A194PYJ5
A0A2H1X2C4 A0A2P8YGX5 A0A3S2LH66 A0A1B6GHB7 A0A2A4JD86 A0A1B6HTG8 A0A2A4IY18 A0A194PXM6 A0A1B6CY04 D2A535 A0A2S2NH85 A0A1B6LYH9 A0A0J7KQN9 A0A1J1HPT6 A0A0K8SJM1 A0A154PAY0 A0A1W4WW74 A0A1Y1KF48 A0A0A9WCX6 A0A2H1WT49 A0A0T6BFY6 A0A2S2PZG0 A0A146LMI6 A0A023F3V3 A0A069DVE7 A0A194QS69 A0A1E1W1C8 A0A2M4AAV7 A0A2M4CTY3 A0A2M4CUE7 A0A2M4BIX1 A0A2M4CSN5 A0A2M4BIV5 A0A2M4CUE3 A0A212FAL1 J9KB87 A0A2H8TM94 Q16W79 A0A2M3Z330 A0A2M3Z363 U4URR0 A0A1Q3FL61 A0A1Q3FKN7 B0WAI1 E2BSZ7 A0A1Q3FLC7 A0A1Q3FLB8 A0A182QCE8 A0A026WMI6 A0A2A4IXD4 A0A182J2E5 A0A0K8VHS8 A0A2R7W706 A0A1L8DE03 A0A182MSS2 A0A182VHT3 A0A2C9H8S5 A0A1Y1KH29 Q5TSW5 A0A1L8DDP9 F4WT02 A0A151ITA2 A0A158NYI1 A0A195AZB4 A0A151WHP4 E2A659 A0A195CVZ7 A0A195F6C4 A0A3S2LGU6 A0A0L7RGJ1 A0A182H1C2 A0A2S2NPL3 A0A084WT83 A0A182N9U1 E9IUT7 A0A194QAI0 H9JAJ5 A0A0M8ZQJ4 A0A212EJ99 A0A182K2U1 A0A2J7PUC7 A0A194RIY8 A0A2H8THB2 A0A182RDY9 A0A182PQS5 A0A182W3L5 A0A0K8WKB4 A0A2W1BZD6 A0A088ANF5 A0A2A3EDD3 D6WRR1 A0A034W601 A0A194PYJ5
EC Number
3.4.-.-
Pubmed
EMBL
KQ460323
KPJ15907.1
KQ459586
KPI98097.1
RSAL01000124
RVE46608.1
+ More
AGBW02009461 OWR50762.1 OWR50760.1 KPJ15909.1 ODYU01012927 SOQ59469.1 PYGN01000603 PSN43506.1 RVE46607.1 GECZ01007935 JAS61834.1 NWSH01001989 PCG69504.1 GECU01029729 JAS77977.1 NWSH01004603 PCG64857.1 KPI98096.1 GEDC01018946 GEDC01008979 JAS18352.1 JAS28319.1 KQ971361 EFA05308.1 GGMR01003880 MBY16499.1 GEBQ01011235 JAT28742.1 LBMM01004254 KMQ92658.1 CVRI01000006 CRK88225.1 GBRD01012475 GBRD01012473 JAG53349.1 KQ434864 KZC08967.1 GEZM01088644 JAV58046.1 GBHO01038000 JAG05604.1 ODYU01010862 SOQ56231.1 LJIG01000707 KRT86233.1 GGMS01001703 MBY70906.1 GDHC01010234 JAQ08395.1 GBBI01002626 JAC16086.1 GBGD01001034 JAC87855.1 KQ461155 KPJ08368.1 GDQN01010353 JAT80701.1 GGFK01004578 MBW37899.1 GGFL01004110 MBW68288.1 GGFL01004789 MBW68967.1 GGFJ01003792 MBW52933.1 GGFL01004111 MBW68289.1 GGFJ01003793 MBW52934.1 GGFL01004788 MBW68966.1 OWR50761.1 ABLF02035671 GFXV01003482 MBW15287.1 CH477573 EAT38831.1 GGFM01002176 MBW22927.1 GGFM01002194 MBW22945.1 KB632410 ERL95208.1 GFDL01006801 JAV28244.1 GFDL01006875 JAV28170.1 DS231872 EDS41404.1 GL450313 EFN81185.1 GFDL01006731 JAV28314.1 GFDL01006678 JAV28367.1 AXCN02001200 KK107151 QOIP01000003 EZA57287.1 RLU24717.1 NWSH01005286 PCG64319.1 GDHF01018012 GDHF01013873 GDHF01003982 JAI34302.1 JAI38441.1 JAI48332.1 KK854400 PTY15462.1 GFDF01009514 JAV04570.1 AXCM01000458 GEZM01088643 GEZM01088642 JAV58047.1 AAAB01008898 EAL40562.4 GFDF01009508 JAV04576.1 GL888331 EGI62615.1 KQ981029 KYN10190.1 ADTU01003883 KQ976701 KYM77299.1 KQ983114 KYQ47337.1 GL437108 EFN71118.1 KQ977220 KYN04851.1 KQ981768 KYN35936.1 RSAL01000011 RVE53552.1 KQ414596 KOC69980.1 JXUM01102960 KQ564767 KXJ71770.1 GGMR01006492 MBY19111.1 ATLV01026865 ATLV01026866 KE525420 KFB53427.1 GL766050 EFZ15659.1 KQ459249 KPJ02439.1 BABH01009475 BABH01009476 BABH01009477 KQ436008 KOX67603.1 AGBW02014503 OWR41557.1 NEVH01021203 PNF19928.1 KPJ15906.1 GFXV01001690 MBW13495.1 GDHF01032155 GDHF01019937 GDHF01013697 GDHF01000723 JAI20159.1 JAI32377.1 JAI38617.1 JAI51591.1 KZ149899 PZC78617.1 KZ288292 PBC29176.1 KQ971354 EFA05964.2 GAKP01009377 JAC49575.1 KPI98098.1
AGBW02009461 OWR50762.1 OWR50760.1 KPJ15909.1 ODYU01012927 SOQ59469.1 PYGN01000603 PSN43506.1 RVE46607.1 GECZ01007935 JAS61834.1 NWSH01001989 PCG69504.1 GECU01029729 JAS77977.1 NWSH01004603 PCG64857.1 KPI98096.1 GEDC01018946 GEDC01008979 JAS18352.1 JAS28319.1 KQ971361 EFA05308.1 GGMR01003880 MBY16499.1 GEBQ01011235 JAT28742.1 LBMM01004254 KMQ92658.1 CVRI01000006 CRK88225.1 GBRD01012475 GBRD01012473 JAG53349.1 KQ434864 KZC08967.1 GEZM01088644 JAV58046.1 GBHO01038000 JAG05604.1 ODYU01010862 SOQ56231.1 LJIG01000707 KRT86233.1 GGMS01001703 MBY70906.1 GDHC01010234 JAQ08395.1 GBBI01002626 JAC16086.1 GBGD01001034 JAC87855.1 KQ461155 KPJ08368.1 GDQN01010353 JAT80701.1 GGFK01004578 MBW37899.1 GGFL01004110 MBW68288.1 GGFL01004789 MBW68967.1 GGFJ01003792 MBW52933.1 GGFL01004111 MBW68289.1 GGFJ01003793 MBW52934.1 GGFL01004788 MBW68966.1 OWR50761.1 ABLF02035671 GFXV01003482 MBW15287.1 CH477573 EAT38831.1 GGFM01002176 MBW22927.1 GGFM01002194 MBW22945.1 KB632410 ERL95208.1 GFDL01006801 JAV28244.1 GFDL01006875 JAV28170.1 DS231872 EDS41404.1 GL450313 EFN81185.1 GFDL01006731 JAV28314.1 GFDL01006678 JAV28367.1 AXCN02001200 KK107151 QOIP01000003 EZA57287.1 RLU24717.1 NWSH01005286 PCG64319.1 GDHF01018012 GDHF01013873 GDHF01003982 JAI34302.1 JAI38441.1 JAI48332.1 KK854400 PTY15462.1 GFDF01009514 JAV04570.1 AXCM01000458 GEZM01088643 GEZM01088642 JAV58047.1 AAAB01008898 EAL40562.4 GFDF01009508 JAV04576.1 GL888331 EGI62615.1 KQ981029 KYN10190.1 ADTU01003883 KQ976701 KYM77299.1 KQ983114 KYQ47337.1 GL437108 EFN71118.1 KQ977220 KYN04851.1 KQ981768 KYN35936.1 RSAL01000011 RVE53552.1 KQ414596 KOC69980.1 JXUM01102960 KQ564767 KXJ71770.1 GGMR01006492 MBY19111.1 ATLV01026865 ATLV01026866 KE525420 KFB53427.1 GL766050 EFZ15659.1 KQ459249 KPJ02439.1 BABH01009475 BABH01009476 BABH01009477 KQ436008 KOX67603.1 AGBW02014503 OWR41557.1 NEVH01021203 PNF19928.1 KPJ15906.1 GFXV01001690 MBW13495.1 GDHF01032155 GDHF01019937 GDHF01013697 GDHF01000723 JAI20159.1 JAI32377.1 JAI38617.1 JAI51591.1 KZ149899 PZC78617.1 KZ288292 PBC29176.1 KQ971354 EFA05964.2 GAKP01009377 JAC49575.1 KPI98098.1
Proteomes
UP000053240
UP000053268
UP000283053
UP000007151
UP000245037
UP000218220
+ More
UP000007266 UP000036403 UP000183832 UP000076502 UP000192223 UP000007819 UP000008820 UP000030742 UP000002320 UP000008237 UP000075886 UP000053097 UP000279307 UP000075880 UP000075883 UP000075903 UP000076407 UP000007062 UP000007755 UP000078492 UP000005205 UP000078540 UP000075809 UP000000311 UP000078542 UP000078541 UP000053825 UP000069940 UP000249989 UP000030765 UP000075884 UP000005204 UP000053105 UP000075881 UP000235965 UP000075900 UP000075885 UP000075920 UP000005203 UP000242457
UP000007266 UP000036403 UP000183832 UP000076502 UP000192223 UP000007819 UP000008820 UP000030742 UP000002320 UP000008237 UP000075886 UP000053097 UP000279307 UP000075880 UP000075883 UP000075903 UP000076407 UP000007062 UP000007755 UP000078492 UP000005205 UP000078540 UP000075809 UP000000311 UP000078542 UP000078541 UP000053825 UP000069940 UP000249989 UP000030765 UP000075884 UP000005204 UP000053105 UP000075881 UP000235965 UP000075900 UP000075885 UP000075920 UP000005203 UP000242457
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A194RE89
A0A194PXP8
A0A3S2PBF1
A0A212FAJ8
A0A212FAK5
A0A194REM1
+ More
A0A2H1X2C4 A0A2P8YGX5 A0A3S2LH66 A0A1B6GHB7 A0A2A4JD86 A0A1B6HTG8 A0A2A4IY18 A0A194PXM6 A0A1B6CY04 D2A535 A0A2S2NH85 A0A1B6LYH9 A0A0J7KQN9 A0A1J1HPT6 A0A0K8SJM1 A0A154PAY0 A0A1W4WW74 A0A1Y1KF48 A0A0A9WCX6 A0A2H1WT49 A0A0T6BFY6 A0A2S2PZG0 A0A146LMI6 A0A023F3V3 A0A069DVE7 A0A194QS69 A0A1E1W1C8 A0A2M4AAV7 A0A2M4CTY3 A0A2M4CUE7 A0A2M4BIX1 A0A2M4CSN5 A0A2M4BIV5 A0A2M4CUE3 A0A212FAL1 J9KB87 A0A2H8TM94 Q16W79 A0A2M3Z330 A0A2M3Z363 U4URR0 A0A1Q3FL61 A0A1Q3FKN7 B0WAI1 E2BSZ7 A0A1Q3FLC7 A0A1Q3FLB8 A0A182QCE8 A0A026WMI6 A0A2A4IXD4 A0A182J2E5 A0A0K8VHS8 A0A2R7W706 A0A1L8DE03 A0A182MSS2 A0A182VHT3 A0A2C9H8S5 A0A1Y1KH29 Q5TSW5 A0A1L8DDP9 F4WT02 A0A151ITA2 A0A158NYI1 A0A195AZB4 A0A151WHP4 E2A659 A0A195CVZ7 A0A195F6C4 A0A3S2LGU6 A0A0L7RGJ1 A0A182H1C2 A0A2S2NPL3 A0A084WT83 A0A182N9U1 E9IUT7 A0A194QAI0 H9JAJ5 A0A0M8ZQJ4 A0A212EJ99 A0A182K2U1 A0A2J7PUC7 A0A194RIY8 A0A2H8THB2 A0A182RDY9 A0A182PQS5 A0A182W3L5 A0A0K8WKB4 A0A2W1BZD6 A0A088ANF5 A0A2A3EDD3 D6WRR1 A0A034W601 A0A194PYJ5
A0A2H1X2C4 A0A2P8YGX5 A0A3S2LH66 A0A1B6GHB7 A0A2A4JD86 A0A1B6HTG8 A0A2A4IY18 A0A194PXM6 A0A1B6CY04 D2A535 A0A2S2NH85 A0A1B6LYH9 A0A0J7KQN9 A0A1J1HPT6 A0A0K8SJM1 A0A154PAY0 A0A1W4WW74 A0A1Y1KF48 A0A0A9WCX6 A0A2H1WT49 A0A0T6BFY6 A0A2S2PZG0 A0A146LMI6 A0A023F3V3 A0A069DVE7 A0A194QS69 A0A1E1W1C8 A0A2M4AAV7 A0A2M4CTY3 A0A2M4CUE7 A0A2M4BIX1 A0A2M4CSN5 A0A2M4BIV5 A0A2M4CUE3 A0A212FAL1 J9KB87 A0A2H8TM94 Q16W79 A0A2M3Z330 A0A2M3Z363 U4URR0 A0A1Q3FL61 A0A1Q3FKN7 B0WAI1 E2BSZ7 A0A1Q3FLC7 A0A1Q3FLB8 A0A182QCE8 A0A026WMI6 A0A2A4IXD4 A0A182J2E5 A0A0K8VHS8 A0A2R7W706 A0A1L8DE03 A0A182MSS2 A0A182VHT3 A0A2C9H8S5 A0A1Y1KH29 Q5TSW5 A0A1L8DDP9 F4WT02 A0A151ITA2 A0A158NYI1 A0A195AZB4 A0A151WHP4 E2A659 A0A195CVZ7 A0A195F6C4 A0A3S2LGU6 A0A0L7RGJ1 A0A182H1C2 A0A2S2NPL3 A0A084WT83 A0A182N9U1 E9IUT7 A0A194QAI0 H9JAJ5 A0A0M8ZQJ4 A0A212EJ99 A0A182K2U1 A0A2J7PUC7 A0A194RIY8 A0A2H8THB2 A0A182RDY9 A0A182PQS5 A0A182W3L5 A0A0K8WKB4 A0A2W1BZD6 A0A088ANF5 A0A2A3EDD3 D6WRR1 A0A034W601 A0A194PYJ5
Ontologies
GO
PANTHER
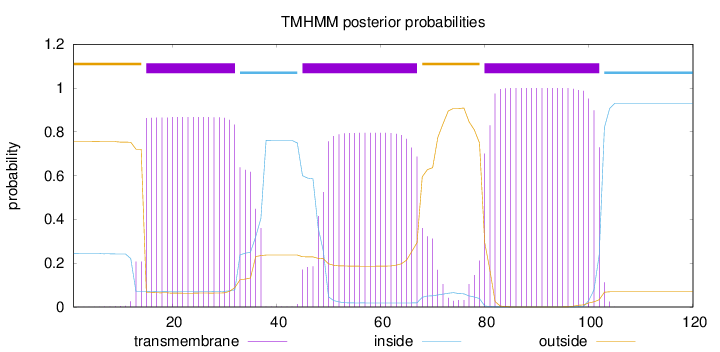
Topology
Length:
120
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
58.28437
Exp number, first 60 AAs:
28.88617
Total prob of N-in:
0.24382
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 32
inside
33 - 44
TMhelix
45 - 67
outside
68 - 79
TMhelix
80 - 102
inside
103 - 120
Population Genetic Test Statistics
Pi
18.678086
Theta
17.905852
Tajima's D
0.031176
CLR
0.882594
CSRT
0.384780760961952
Interpretation
Uncertain
