Gene
KWMTBOMO13387
Pre Gene Modal
BGIBMGA010935
Annotation
PREDICTED:_proton-coupled_folate_transporter-like?_partial_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.686
Sequence
CDS
ATGGCCACAACACTAAATAAAGAAATATCTATACAAGAATCAGAAGCTCAGAACACTCCGAACAATGTTGCAATCAAGAAGAAAACTGGCTTTAGGTACTTACTGGGCATTATTACAGTAGAACCAGTGGCGTTCATAGTACTGTTCGCAGGTCTACTGTCAGGTATGTCGACACAGAATTTAGGCTTAGAAAAAGCTTGTAGAGTAAGTCTTAAACTCGAAGATGCTATCTGTGATTCACTGAGAGCACAAAACCAATCGAATAACTTCACTGATTATGAGAGAATGGTCCAAGAGCATTATTCTGCCCAGCTAAGGTGGAAAAGCGCTTTGCAGTCCGTATTACCGCTCCCTATGCTGCTGTTTGCTGGAGGCTGGAGCGACAAGACAGGCAGAAGAAAAGCCATCATTGTAGTTCCCATAATCGGCGAAACTCTCCAATGCATCAGCAACATCTTAAATGTGGTATTCTTCTACGAAATCCCGCTTGAGGCTTTGATATTCTTTGATACATTCTTCTCGGGGATCACTGGGGGCCAAATCATAATGCTCTTAGCGCTGTTCAATTATATCTGCGATATAACTACCTCGGAGAACAGAACTCAGAGGTTGGGAATCGTTAATGTGTGTACGTTTGTCGGAACGCCGCTAGGTTTGGCGTTATCCGGGATAATGTTGAGGCATCTTGGGTACTATGCCATATATGGAACAGCTCTTTTTTTGCACTTTATAAACTTTTTGTATGTGACATTCAGGATAAAGGACCCATTGAGGACTAAAGAGCAAATTATGACGGTGAGTAATTATTTCTTTTCACTTCTCGTTAATAACATACCGCTGGAATAG
Protein
MATTLNKEISIQESEAQNTPNNVAIKKKTGFRYLLGIITVEPVAFIVLFAGLLSGMSTQNLGLEKACRVSLKLEDAICDSLRAQNQSNNFTDYERMVQEHYSAQLRWKSALQSVLPLPMLLFAGGWSDKTGRRKAIIVVPIIGETLQCISNILNVVFFYEIPLEALIFFDTFFSGITGGQIIMLLALFNYICDITTSENRTQRLGIVNVCTFVGTPLGLALSGIMLRHLGYYAIYGTALFLHFINFLYVTFRIKDPLRTKEQIMTVSNYFFSLLVNNIPLE
Summary
Uniprot
H9JN32
H9JN33
A0A0L7LL21
A0A2H1W9L1
A0A0L7KYE5
H9JN31
+ More
A0A2A4IW55 A0A2J7PUB6 A0A2H1WQK1 H9JN07 A0A0L7KUI5 A0A212FAL1 A0A2H1V1J8 A0A2A4ISH2 A0A194PXN1 A0A194RDI4 A0A212FAK5 A0A194REM1 A0A2H1WVH5 A0A212EU49 A0A0L7L9Z5 A0A2A4IXD4 A0A2A4IY18 A0A194PXP8 A0A194RE89 A0A2P8YGX7 A0A194PZD1 A0A194PYJ5 A0A3S2LLF7 A0A0L7KST7 A0A194RF29 Q16W79 A0A3S2PBF1 A0A067RJA9 A0A194QSS2 A0A212FAJ8 A0A0L7L4T7 A0A194QAI0 A0A3S2PEP4 A0A224XHS1 A0A1B6LYH9 A0A1B6IHD1 A0A194Q3W9 A0A1L8DDP9 A0A212FAM1 T1HEN2 A0A194RDJ0 A0A2A4J4Z4 H9JN34 H9JN35 A0A2H1W938 A0A1B0CBZ1 A0A1Q3FLB8 A0A182QCE8 A0A1Q3FLC7 A0A2R7WWJ8 B0WAI1 A0A2M4AAV7 A0A182U4U1 H9JN30 A0A182MSS2 H9JAJ5 A0A2H1WJM3 A0A2M4CUE7 A0A1W4WW74 A0A2M4CSN5 A0A2M4CTY3 A0A2M4BIX1 A0A2M4BIV5 A0A3Q0IJ06 A0A1A9XH14 A0A2H1WT49 A0A2M3Z363 A0A2M3Z330 A0A1B0ANC8 A0A182VHT3 Q5TSW5 A0A2M4CUE3 A0A2W1BZD6 A0A1B0FMN2 A0A1A9WCM0 A0A069DVE7 A0A182J2E5 A0A2C9H8S5 A0A3S2LGU6 A0A212EU39 B4R4V6 A0A023F3V3 B3NUZ2 A0A2S2PXN1 A0A1W4VHL4 A0A0L7LPQ8 A0A1A9ZI85 Q9VY22 A0A1Y1KH29 Q6NPA2 A0A1J1HPT6 A0A3B0K378 B4PW63 A0A0A1XR69
A0A2A4IW55 A0A2J7PUB6 A0A2H1WQK1 H9JN07 A0A0L7KUI5 A0A212FAL1 A0A2H1V1J8 A0A2A4ISH2 A0A194PXN1 A0A194RDI4 A0A212FAK5 A0A194REM1 A0A2H1WVH5 A0A212EU49 A0A0L7L9Z5 A0A2A4IXD4 A0A2A4IY18 A0A194PXP8 A0A194RE89 A0A2P8YGX7 A0A194PZD1 A0A194PYJ5 A0A3S2LLF7 A0A0L7KST7 A0A194RF29 Q16W79 A0A3S2PBF1 A0A067RJA9 A0A194QSS2 A0A212FAJ8 A0A0L7L4T7 A0A194QAI0 A0A3S2PEP4 A0A224XHS1 A0A1B6LYH9 A0A1B6IHD1 A0A194Q3W9 A0A1L8DDP9 A0A212FAM1 T1HEN2 A0A194RDJ0 A0A2A4J4Z4 H9JN34 H9JN35 A0A2H1W938 A0A1B0CBZ1 A0A1Q3FLB8 A0A182QCE8 A0A1Q3FLC7 A0A2R7WWJ8 B0WAI1 A0A2M4AAV7 A0A182U4U1 H9JN30 A0A182MSS2 H9JAJ5 A0A2H1WJM3 A0A2M4CUE7 A0A1W4WW74 A0A2M4CSN5 A0A2M4CTY3 A0A2M4BIX1 A0A2M4BIV5 A0A3Q0IJ06 A0A1A9XH14 A0A2H1WT49 A0A2M3Z363 A0A2M3Z330 A0A1B0ANC8 A0A182VHT3 Q5TSW5 A0A2M4CUE3 A0A2W1BZD6 A0A1B0FMN2 A0A1A9WCM0 A0A069DVE7 A0A182J2E5 A0A2C9H8S5 A0A3S2LGU6 A0A212EU39 B4R4V6 A0A023F3V3 B3NUZ2 A0A2S2PXN1 A0A1W4VHL4 A0A0L7LPQ8 A0A1A9ZI85 Q9VY22 A0A1Y1KH29 Q6NPA2 A0A1J1HPT6 A0A3B0K378 B4PW63 A0A0A1XR69
Pubmed
EMBL
BABH01030455
BABH01030456
BABH01030448
JTDY01000713
KOB76125.1
ODYU01007183
+ More
SOQ49737.1 JTDY01004446 KOB68155.1 BABH01030471 NWSH01006054 PCG63706.1 NEVH01021203 PNF19923.1 ODYU01010298 SOQ55308.1 BABH01041390 BABH01041391 BABH01041392 JTDY01005733 KOB66684.1 AGBW02009461 OWR50761.1 ODYU01000073 SOQ34212.1 NWSH01007506 PCG62937.1 KQ459586 KPI98101.1 KQ460323 KPJ15903.1 OWR50760.1 KPJ15909.1 ODYU01011363 SOQ57061.1 AGBW02012463 OWR45016.1 JTDY01002065 KOB72195.1 NWSH01005286 PCG64319.1 NWSH01004603 PCG64857.1 KPI98097.1 KPJ15907.1 PYGN01000603 PSN43505.1 KPI98099.1 KPI98098.1 RSAL01000068 RVE49265.1 JTDY01006280 KOB66145.1 KPJ15905.1 CH477573 EAT38831.1 RSAL01000124 RVE46608.1 KK852651 KDR19441.1 KQ461155 KPJ08369.1 OWR50762.1 JTDY01002998 KOB70346.1 KQ459249 KPJ02439.1 RVE49267.1 GFTR01004389 JAW12037.1 GEBQ01011235 JAT28742.1 GECU01021378 JAS86328.1 KPI98100.1 GFDF01009508 JAV04576.1 OWR50763.1 ACPB03012810 KPJ15908.1 NWSH01002978 PCG67155.1 BABH01030450 ODYU01007104 SOQ49580.1 AJWK01006000 AJWK01006001 GFDL01006678 JAV28367.1 AXCN02001200 GFDL01006731 JAV28314.1 KK855798 PTY23912.1 DS231872 EDS41404.1 GGFK01004578 MBW37899.1 BABH01030475 AXCM01000458 BABH01009475 BABH01009476 BABH01009477 ODYU01009084 SOQ53249.1 GGFL01004789 MBW68967.1 GGFL01004111 MBW68289.1 GGFL01004110 MBW68288.1 GGFJ01003792 MBW52933.1 GGFJ01003793 MBW52934.1 ODYU01010862 SOQ56231.1 GGFM01002194 MBW22945.1 GGFM01002176 MBW22927.1 JXJN01000793 AAAB01008898 EAL40562.4 GGFL01004788 MBW68966.1 KZ149899 PZC78617.1 CCAG010001068 GBGD01001034 JAC87855.1 RSAL01000011 RVE53552.1 OWR45015.1 CM000366 EDX17913.1 GBBI01002626 JAC16086.1 CH954180 EDV47090.1 GGMS01000917 MBY70120.1 JTDY01000388 KOB77422.1 AE014298 BT133123 AAF48383.1 AEY64291.1 GEZM01088643 GEZM01088642 JAV58047.1 BT011024 AAR30184.1 CVRI01000006 CRK88225.1 OUUW01000015 SPP88717.1 CM000162 EDX01694.1 KRK06299.1 GBXI01016992 GBXI01013508 GBXI01006853 GBXI01003332 GBXI01000927 JAC97299.1 JAD00784.1 JAD07439.1 JAD10960.1 JAD13365.1
SOQ49737.1 JTDY01004446 KOB68155.1 BABH01030471 NWSH01006054 PCG63706.1 NEVH01021203 PNF19923.1 ODYU01010298 SOQ55308.1 BABH01041390 BABH01041391 BABH01041392 JTDY01005733 KOB66684.1 AGBW02009461 OWR50761.1 ODYU01000073 SOQ34212.1 NWSH01007506 PCG62937.1 KQ459586 KPI98101.1 KQ460323 KPJ15903.1 OWR50760.1 KPJ15909.1 ODYU01011363 SOQ57061.1 AGBW02012463 OWR45016.1 JTDY01002065 KOB72195.1 NWSH01005286 PCG64319.1 NWSH01004603 PCG64857.1 KPI98097.1 KPJ15907.1 PYGN01000603 PSN43505.1 KPI98099.1 KPI98098.1 RSAL01000068 RVE49265.1 JTDY01006280 KOB66145.1 KPJ15905.1 CH477573 EAT38831.1 RSAL01000124 RVE46608.1 KK852651 KDR19441.1 KQ461155 KPJ08369.1 OWR50762.1 JTDY01002998 KOB70346.1 KQ459249 KPJ02439.1 RVE49267.1 GFTR01004389 JAW12037.1 GEBQ01011235 JAT28742.1 GECU01021378 JAS86328.1 KPI98100.1 GFDF01009508 JAV04576.1 OWR50763.1 ACPB03012810 KPJ15908.1 NWSH01002978 PCG67155.1 BABH01030450 ODYU01007104 SOQ49580.1 AJWK01006000 AJWK01006001 GFDL01006678 JAV28367.1 AXCN02001200 GFDL01006731 JAV28314.1 KK855798 PTY23912.1 DS231872 EDS41404.1 GGFK01004578 MBW37899.1 BABH01030475 AXCM01000458 BABH01009475 BABH01009476 BABH01009477 ODYU01009084 SOQ53249.1 GGFL01004789 MBW68967.1 GGFL01004111 MBW68289.1 GGFL01004110 MBW68288.1 GGFJ01003792 MBW52933.1 GGFJ01003793 MBW52934.1 ODYU01010862 SOQ56231.1 GGFM01002194 MBW22945.1 GGFM01002176 MBW22927.1 JXJN01000793 AAAB01008898 EAL40562.4 GGFL01004788 MBW68966.1 KZ149899 PZC78617.1 CCAG010001068 GBGD01001034 JAC87855.1 RSAL01000011 RVE53552.1 OWR45015.1 CM000366 EDX17913.1 GBBI01002626 JAC16086.1 CH954180 EDV47090.1 GGMS01000917 MBY70120.1 JTDY01000388 KOB77422.1 AE014298 BT133123 AAF48383.1 AEY64291.1 GEZM01088643 GEZM01088642 JAV58047.1 BT011024 AAR30184.1 CVRI01000006 CRK88225.1 OUUW01000015 SPP88717.1 CM000162 EDX01694.1 KRK06299.1 GBXI01016992 GBXI01013508 GBXI01006853 GBXI01003332 GBXI01000927 JAC97299.1 JAD00784.1 JAD07439.1 JAD10960.1 JAD13365.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000235965
UP000007151
UP000053268
+ More
UP000053240 UP000245037 UP000283053 UP000008820 UP000027135 UP000015103 UP000092461 UP000075886 UP000002320 UP000075902 UP000075883 UP000192223 UP000079169 UP000092443 UP000092460 UP000075903 UP000007062 UP000092444 UP000091820 UP000075880 UP000076407 UP000000304 UP000008711 UP000192221 UP000092445 UP000000803 UP000183832 UP000268350 UP000002282
UP000053240 UP000245037 UP000283053 UP000008820 UP000027135 UP000015103 UP000092461 UP000075886 UP000002320 UP000075902 UP000075883 UP000192223 UP000079169 UP000092443 UP000092460 UP000075903 UP000007062 UP000092444 UP000091820 UP000075880 UP000076407 UP000000304 UP000008711 UP000192221 UP000092445 UP000000803 UP000183832 UP000268350 UP000002282
Pfam
PF07690 MFS_1
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JN32
H9JN33
A0A0L7LL21
A0A2H1W9L1
A0A0L7KYE5
H9JN31
+ More
A0A2A4IW55 A0A2J7PUB6 A0A2H1WQK1 H9JN07 A0A0L7KUI5 A0A212FAL1 A0A2H1V1J8 A0A2A4ISH2 A0A194PXN1 A0A194RDI4 A0A212FAK5 A0A194REM1 A0A2H1WVH5 A0A212EU49 A0A0L7L9Z5 A0A2A4IXD4 A0A2A4IY18 A0A194PXP8 A0A194RE89 A0A2P8YGX7 A0A194PZD1 A0A194PYJ5 A0A3S2LLF7 A0A0L7KST7 A0A194RF29 Q16W79 A0A3S2PBF1 A0A067RJA9 A0A194QSS2 A0A212FAJ8 A0A0L7L4T7 A0A194QAI0 A0A3S2PEP4 A0A224XHS1 A0A1B6LYH9 A0A1B6IHD1 A0A194Q3W9 A0A1L8DDP9 A0A212FAM1 T1HEN2 A0A194RDJ0 A0A2A4J4Z4 H9JN34 H9JN35 A0A2H1W938 A0A1B0CBZ1 A0A1Q3FLB8 A0A182QCE8 A0A1Q3FLC7 A0A2R7WWJ8 B0WAI1 A0A2M4AAV7 A0A182U4U1 H9JN30 A0A182MSS2 H9JAJ5 A0A2H1WJM3 A0A2M4CUE7 A0A1W4WW74 A0A2M4CSN5 A0A2M4CTY3 A0A2M4BIX1 A0A2M4BIV5 A0A3Q0IJ06 A0A1A9XH14 A0A2H1WT49 A0A2M3Z363 A0A2M3Z330 A0A1B0ANC8 A0A182VHT3 Q5TSW5 A0A2M4CUE3 A0A2W1BZD6 A0A1B0FMN2 A0A1A9WCM0 A0A069DVE7 A0A182J2E5 A0A2C9H8S5 A0A3S2LGU6 A0A212EU39 B4R4V6 A0A023F3V3 B3NUZ2 A0A2S2PXN1 A0A1W4VHL4 A0A0L7LPQ8 A0A1A9ZI85 Q9VY22 A0A1Y1KH29 Q6NPA2 A0A1J1HPT6 A0A3B0K378 B4PW63 A0A0A1XR69
A0A2A4IW55 A0A2J7PUB6 A0A2H1WQK1 H9JN07 A0A0L7KUI5 A0A212FAL1 A0A2H1V1J8 A0A2A4ISH2 A0A194PXN1 A0A194RDI4 A0A212FAK5 A0A194REM1 A0A2H1WVH5 A0A212EU49 A0A0L7L9Z5 A0A2A4IXD4 A0A2A4IY18 A0A194PXP8 A0A194RE89 A0A2P8YGX7 A0A194PZD1 A0A194PYJ5 A0A3S2LLF7 A0A0L7KST7 A0A194RF29 Q16W79 A0A3S2PBF1 A0A067RJA9 A0A194QSS2 A0A212FAJ8 A0A0L7L4T7 A0A194QAI0 A0A3S2PEP4 A0A224XHS1 A0A1B6LYH9 A0A1B6IHD1 A0A194Q3W9 A0A1L8DDP9 A0A212FAM1 T1HEN2 A0A194RDJ0 A0A2A4J4Z4 H9JN34 H9JN35 A0A2H1W938 A0A1B0CBZ1 A0A1Q3FLB8 A0A182QCE8 A0A1Q3FLC7 A0A2R7WWJ8 B0WAI1 A0A2M4AAV7 A0A182U4U1 H9JN30 A0A182MSS2 H9JAJ5 A0A2H1WJM3 A0A2M4CUE7 A0A1W4WW74 A0A2M4CSN5 A0A2M4CTY3 A0A2M4BIX1 A0A2M4BIV5 A0A3Q0IJ06 A0A1A9XH14 A0A2H1WT49 A0A2M3Z363 A0A2M3Z330 A0A1B0ANC8 A0A182VHT3 Q5TSW5 A0A2M4CUE3 A0A2W1BZD6 A0A1B0FMN2 A0A1A9WCM0 A0A069DVE7 A0A182J2E5 A0A2C9H8S5 A0A3S2LGU6 A0A212EU39 B4R4V6 A0A023F3V3 B3NUZ2 A0A2S2PXN1 A0A1W4VHL4 A0A0L7LPQ8 A0A1A9ZI85 Q9VY22 A0A1Y1KH29 Q6NPA2 A0A1J1HPT6 A0A3B0K378 B4PW63 A0A0A1XR69
Ontologies
GO
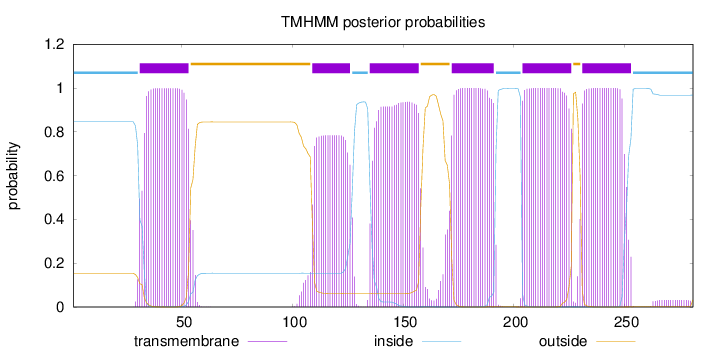
Topology
Length:
281
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
124.36659
Exp number, first 60 AAs:
22.55046
Total prob of N-in:
0.84624
POSSIBLE N-term signal
sequence
inside
1 - 30
TMhelix
31 - 53
outside
54 - 108
TMhelix
109 - 126
inside
127 - 134
TMhelix
135 - 157
outside
158 - 171
TMhelix
172 - 191
inside
192 - 203
TMhelix
204 - 226
outside
227 - 230
TMhelix
231 - 253
inside
254 - 281
Population Genetic Test Statistics
Pi
15.029232
Theta
21.349273
Tajima's D
-0.779262
CLR
39.571505
CSRT
0.182240887955602
Interpretation
Uncertain
