Gene
KWMTBOMO13386
Annotation
PREDICTED:_solute_carrier_family_46_member_3-like_isoform_X3_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.91
Sequence
CDS
ATGACAGGCGGTAATCAAGAAAATTTAAATGAAAAACAGGAAGATGAATTCACTGAAGATGATCCATTAAGAAGCCAAGATGTTAAGGAAGTCTCACAACCAAGCGTATGTGCTAAATTCTGCCATCTAATCAAAAACATGACAGTGGAAGCAAGTTTGTTATTCCTCATCATGTCCAGTCTTATTTCAATGTTGACAACGCAAAACTTAAGCTTAGAAAAAGCTTGCCGAGTGAACTTAAATTTTTCAGATGAAATCTGCACCTCCCTAAAGCTGCAAGATTCTGATGTCAACAGTCAGAATCAGTATGAAAAAGAGACTCAGAAACTGCTAGCGTCTGTAATGGCTAAAAGAACTTATATTTCCGCCACGATACCATGCGTGATCGCCTTATTAGTCGGAGCATGGTCCGACAAAACTGGACACAGGAAAGTGTTTCTGACAATTTCAATATTTGGGCAATTCCTGATTACAATTAATAATATGTTAAATGTTTATTTCTTTCACGAGCTAAGCGTAGAAGTTCTCGTGTTCAGTGAAGCTATTTTAGAAAGTTTTTCGGGTGGTTGGTGCGTAGGTATCTTAACTGCCTTTTCATATATCTCTGCAATTACATCCGATGAAGACCGTACTTACAGAATGGGATTGTTAAACTTCTGTATGACAGTTGGATTTCCGGTTGGTATGGGACTCAGCGGCATACTCTTGAAGAAATACGGTTATTACGGATCTTACGGACTAGCTGGTGCAATTCACAGCTTGAATGTTATGTACAGCATATTTGTATTGAAGGATCCAAAGAGAAATAAAGAACAAAAGAAGTACGATAAACAAGGAATACGCCACTTTTTTCGAACCTTCTTCGATTTTGCCAACGTTAAGGATACAGTAGGTGTTGTCTTCAAGAAAGCACCGAACAACAGAAGAGTCCGTCTTTGTATTCTTCTGGCAGTAGTTACAGTTTTATTCGGACCAATGTATGGTGAAGTATCTATATTATATTTGTCGACCCGCTACCGATTCAACTGGGATGAAATCAAATTCAGTATTTTCCAGACATACAACTTTGTAACGCACACAGTTGGTACAATTTTCTCGATATTATTCTTTAGCAAATACCTCAAATGGGACGACTCAATATTGGGCATAATATCGACGATCAGTAAAATAGCGGGATCTTTCGTCTACTGTTTTGCACCGAACGATAAAATATTCTTTATTGCGCCTCTGGTTGAAATATTGAATGGAACAGCGCTTTTGGCAATGAGGTCTATATTCTCCAAGATGGTGCAGCCGGATGAACTCGGTAAAATGAACTCAGTCGTGGGCTTGACCGAGAATCTGATGCCATTACTATACGTACCGCTCTACACTAAAGTCTACACGTCCACAATGGAAGTTCTACCGGGGGCCGTGTTTCTGATGGGCGCTCTCATGACTCTACCAGCTGTTGGAGCTTTCATATGGCTATTTGTCGAACACAGGCTAAGTCTAAGGAAAACGAAATCCGAAATCGAAAATCAATCTCCTGATTAG
Protein
MTGGNQENLNEKQEDEFTEDDPLRSQDVKEVSQPSVCAKFCHLIKNMTVEASLLFLIMSSLISMLTTQNLSLEKACRVNLNFSDEICTSLKLQDSDVNSQNQYEKETQKLLASVMAKRTYISATIPCVIALLVGAWSDKTGHRKVFLTISIFGQFLITINNMLNVYFFHELSVEVLVFSEAILESFSGGWCVGILTAFSYISAITSDEDRTYRMGLLNFCMTVGFPVGMGLSGILLKKYGYYGSYGLAGAIHSLNVMYSIFVLKDPKRNKEQKKYDKQGIRHFFRTFFDFANVKDTVGVVFKKAPNNRRVRLCILLAVVTVLFGPMYGEVSILYLSTRYRFNWDEIKFSIFQTYNFVTHTVGTIFSILFFSKYLKWDDSILGIISTISKIAGSFVYCFAPNDKIFFIAPLVEILNGTALLAMRSIFSKMVQPDELGKMNSVVGLTENLMPLLYVPLYTKVYTSTMEVLPGAVFLMGALMTLPAVGAFIWLFVEHRLSLRKTKSEIENQSPD
Summary
Uniprot
A0A2H1WT49
A0A3S2LLF7
A0A2A4IXD4
A0A194PYJ5
A0A212FAM1
A0A0L7LL21
+ More
A0A212FAL1 A0A194PXP8 A0A194RE89 A0A3S2PBF1 A0A194RDJ0 A0A212FAJ8 A0A212FAK5 A0A194PXM6 A0A194REM1 A0A2A4IY18 A0A3S2LGU6 A0A2W1BZD6 A0A146LMI6 A0A023F3V3 A0A069DVE7 H9JAJ5 A0A2H1W938 A0A1J1HPT6 A0A194QAI0 A0A154PAY0 A0A0L7RGJ1 K7J2K2 A0A088ANF5 A0A1I8M3Q0 Q5TSW5 A0A1B6CY04 A0A2M3Z363 A0A2M4BIV5 A0A2M4BIX1 A0A2M3Z330 A0A1B6LYH9 D6WRR1 A0A2C9H8S5 A0A182MSS2 A0A182VHT3 A0A2M4CUE7 A0A2M4AAV7 A0A2M4CSN5 A0A2M4CUE3 A0A2M4CTY3 A0A182QCE8 A0A182J2E5 A0A212EJ99 A0A212EU49 A0A1Y1KH29 A0A067RJA9 A0A2A3EDD3 A0A0M8ZQJ4 Q16W79 A0A2S2R688 J9KB87 A0A2H8TM94 A0A1W4WW74 A0A1I8PUY1 W8C3P0 A0A1L8DDP9 A0A1Q3FLB8 A0A1Q3FLC7 E9IUT7 B0WAI1 A0A026WMI6 A0A194RDI4 A0A034W601 A0A158NYI1 A0A151ITA2 D2A535 E2A659 F4WT02 A0A0L0C7Q4 A0A195F6C4 A0A195AZB4 A0A195CVZ7 A0A194PXN1 A0A0K8WKB4 U4URR0 A0A2H1WJM3 A0A151WHP4 A0A2S2PEA7 Q6NPA2 A0A194Q3W9 E2BSZ7 A0A2A4IW55 A0A2H1V1J8 A0A0A1XR69 B4R4V6 A0A2H8TQH7 B4PW63 Q9VY22 B4L4F7 A0A1W4XEM7 X1WII3 B4M2P7 A0A1B6HTG8 B3NUZ2
A0A212FAL1 A0A194PXP8 A0A194RE89 A0A3S2PBF1 A0A194RDJ0 A0A212FAJ8 A0A212FAK5 A0A194PXM6 A0A194REM1 A0A2A4IY18 A0A3S2LGU6 A0A2W1BZD6 A0A146LMI6 A0A023F3V3 A0A069DVE7 H9JAJ5 A0A2H1W938 A0A1J1HPT6 A0A194QAI0 A0A154PAY0 A0A0L7RGJ1 K7J2K2 A0A088ANF5 A0A1I8M3Q0 Q5TSW5 A0A1B6CY04 A0A2M3Z363 A0A2M4BIV5 A0A2M4BIX1 A0A2M3Z330 A0A1B6LYH9 D6WRR1 A0A2C9H8S5 A0A182MSS2 A0A182VHT3 A0A2M4CUE7 A0A2M4AAV7 A0A2M4CSN5 A0A2M4CUE3 A0A2M4CTY3 A0A182QCE8 A0A182J2E5 A0A212EJ99 A0A212EU49 A0A1Y1KH29 A0A067RJA9 A0A2A3EDD3 A0A0M8ZQJ4 Q16W79 A0A2S2R688 J9KB87 A0A2H8TM94 A0A1W4WW74 A0A1I8PUY1 W8C3P0 A0A1L8DDP9 A0A1Q3FLB8 A0A1Q3FLC7 E9IUT7 B0WAI1 A0A026WMI6 A0A194RDI4 A0A034W601 A0A158NYI1 A0A151ITA2 D2A535 E2A659 F4WT02 A0A0L0C7Q4 A0A195F6C4 A0A195AZB4 A0A195CVZ7 A0A194PXN1 A0A0K8WKB4 U4URR0 A0A2H1WJM3 A0A151WHP4 A0A2S2PEA7 Q6NPA2 A0A194Q3W9 E2BSZ7 A0A2A4IW55 A0A2H1V1J8 A0A0A1XR69 B4R4V6 A0A2H8TQH7 B4PW63 Q9VY22 B4L4F7 A0A1W4XEM7 X1WII3 B4M2P7 A0A1B6HTG8 B3NUZ2
Pubmed
26354079
22118469
26227816
28756777
26823975
25474469
+ More
26334808 19121390 20075255 25315136 12364791 14747013 17210077 18362917 19820115 28004739 24845553 17510324 24495485 21282665 24508170 30249741 25348373 21347285 20798317 21719571 26108605 23537049 25830018 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
26334808 19121390 20075255 25315136 12364791 14747013 17210077 18362917 19820115 28004739 24845553 17510324 24495485 21282665 24508170 30249741 25348373 21347285 20798317 21719571 26108605 23537049 25830018 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
ODYU01010862
SOQ56231.1
RSAL01000068
RVE49265.1
NWSH01005286
PCG64319.1
+ More
KQ459586 KPI98098.1 AGBW02009461 OWR50763.1 JTDY01000713 KOB76125.1 OWR50761.1 KPI98097.1 KQ460323 KPJ15907.1 RSAL01000124 RVE46608.1 KPJ15908.1 OWR50762.1 OWR50760.1 KPI98096.1 KPJ15909.1 NWSH01004603 PCG64857.1 RSAL01000011 RVE53552.1 KZ149899 PZC78617.1 GDHC01010234 JAQ08395.1 GBBI01002626 JAC16086.1 GBGD01001034 JAC87855.1 BABH01009475 BABH01009476 BABH01009477 ODYU01007104 SOQ49580.1 CVRI01000006 CRK88225.1 KQ459249 KPJ02439.1 KQ434864 KZC08967.1 KQ414596 KOC69980.1 AAAB01008898 EAL40562.4 GEDC01018946 GEDC01008979 JAS18352.1 JAS28319.1 GGFM01002194 MBW22945.1 GGFJ01003793 MBW52934.1 GGFJ01003792 MBW52933.1 GGFM01002176 MBW22927.1 GEBQ01011235 JAT28742.1 KQ971354 EFA05964.2 AXCM01000458 GGFL01004789 MBW68967.1 GGFK01004578 MBW37899.1 GGFL01004111 MBW68289.1 GGFL01004788 MBW68966.1 GGFL01004110 MBW68288.1 AXCN02001200 AGBW02014503 OWR41557.1 AGBW02012463 OWR45016.1 GEZM01088643 GEZM01088642 JAV58047.1 KK852651 KDR19441.1 KZ288292 PBC29176.1 KQ436008 KOX67603.1 CH477573 EAT38831.1 GGMS01016275 MBY85478.1 ABLF02035671 GFXV01003482 MBW15287.1 GAMC01005989 JAC00567.1 GFDF01009508 JAV04576.1 GFDL01006678 JAV28367.1 GFDL01006731 JAV28314.1 GL766050 EFZ15659.1 DS231872 EDS41404.1 KK107151 QOIP01000003 EZA57287.1 RLU24717.1 KPJ15903.1 GAKP01009377 JAC49575.1 ADTU01003883 KQ981029 KYN10190.1 KQ971361 EFA05308.1 GL437108 EFN71118.1 GL888331 EGI62615.1 JRES01000902 KNC27439.1 KQ981768 KYN35936.1 KQ976701 KYM77299.1 KQ977220 KYN04851.1 KPI98101.1 GDHF01032155 GDHF01019937 GDHF01013697 GDHF01000723 JAI20159.1 JAI32377.1 JAI38617.1 JAI51591.1 KB632410 ERL95208.1 ODYU01009084 SOQ53249.1 KQ983114 KYQ47337.1 GGMR01015172 MBY27791.1 BT011024 AAR30184.1 KPI98100.1 GL450313 EFN81185.1 NWSH01006054 PCG63706.1 ODYU01000073 SOQ34212.1 GBXI01016992 GBXI01013508 GBXI01006853 GBXI01003332 GBXI01000927 JAC97299.1 JAD00784.1 JAD07439.1 JAD10960.1 JAD13365.1 CM000366 EDX17913.1 GFXV01004631 MBW16436.1 CM000162 EDX01694.1 KRK06299.1 AE014298 BT133123 AAF48383.1 AEY64291.1 CH933810 EDW07435.1 ABLF02035661 CH940651 EDW65951.1 GECU01029729 JAS77977.1 CH954180 EDV47090.1
KQ459586 KPI98098.1 AGBW02009461 OWR50763.1 JTDY01000713 KOB76125.1 OWR50761.1 KPI98097.1 KQ460323 KPJ15907.1 RSAL01000124 RVE46608.1 KPJ15908.1 OWR50762.1 OWR50760.1 KPI98096.1 KPJ15909.1 NWSH01004603 PCG64857.1 RSAL01000011 RVE53552.1 KZ149899 PZC78617.1 GDHC01010234 JAQ08395.1 GBBI01002626 JAC16086.1 GBGD01001034 JAC87855.1 BABH01009475 BABH01009476 BABH01009477 ODYU01007104 SOQ49580.1 CVRI01000006 CRK88225.1 KQ459249 KPJ02439.1 KQ434864 KZC08967.1 KQ414596 KOC69980.1 AAAB01008898 EAL40562.4 GEDC01018946 GEDC01008979 JAS18352.1 JAS28319.1 GGFM01002194 MBW22945.1 GGFJ01003793 MBW52934.1 GGFJ01003792 MBW52933.1 GGFM01002176 MBW22927.1 GEBQ01011235 JAT28742.1 KQ971354 EFA05964.2 AXCM01000458 GGFL01004789 MBW68967.1 GGFK01004578 MBW37899.1 GGFL01004111 MBW68289.1 GGFL01004788 MBW68966.1 GGFL01004110 MBW68288.1 AXCN02001200 AGBW02014503 OWR41557.1 AGBW02012463 OWR45016.1 GEZM01088643 GEZM01088642 JAV58047.1 KK852651 KDR19441.1 KZ288292 PBC29176.1 KQ436008 KOX67603.1 CH477573 EAT38831.1 GGMS01016275 MBY85478.1 ABLF02035671 GFXV01003482 MBW15287.1 GAMC01005989 JAC00567.1 GFDF01009508 JAV04576.1 GFDL01006678 JAV28367.1 GFDL01006731 JAV28314.1 GL766050 EFZ15659.1 DS231872 EDS41404.1 KK107151 QOIP01000003 EZA57287.1 RLU24717.1 KPJ15903.1 GAKP01009377 JAC49575.1 ADTU01003883 KQ981029 KYN10190.1 KQ971361 EFA05308.1 GL437108 EFN71118.1 GL888331 EGI62615.1 JRES01000902 KNC27439.1 KQ981768 KYN35936.1 KQ976701 KYM77299.1 KQ977220 KYN04851.1 KPI98101.1 GDHF01032155 GDHF01019937 GDHF01013697 GDHF01000723 JAI20159.1 JAI32377.1 JAI38617.1 JAI51591.1 KB632410 ERL95208.1 ODYU01009084 SOQ53249.1 KQ983114 KYQ47337.1 GGMR01015172 MBY27791.1 BT011024 AAR30184.1 KPI98100.1 GL450313 EFN81185.1 NWSH01006054 PCG63706.1 ODYU01000073 SOQ34212.1 GBXI01016992 GBXI01013508 GBXI01006853 GBXI01003332 GBXI01000927 JAC97299.1 JAD00784.1 JAD07439.1 JAD10960.1 JAD13365.1 CM000366 EDX17913.1 GFXV01004631 MBW16436.1 CM000162 EDX01694.1 KRK06299.1 AE014298 BT133123 AAF48383.1 AEY64291.1 CH933810 EDW07435.1 ABLF02035661 CH940651 EDW65951.1 GECU01029729 JAS77977.1 CH954180 EDV47090.1
Proteomes
UP000283053
UP000218220
UP000053268
UP000007151
UP000037510
UP000053240
+ More
UP000005204 UP000183832 UP000076502 UP000053825 UP000002358 UP000005203 UP000095301 UP000007062 UP000007266 UP000076407 UP000075883 UP000075903 UP000075886 UP000075880 UP000027135 UP000242457 UP000053105 UP000008820 UP000007819 UP000192223 UP000095300 UP000002320 UP000053097 UP000279307 UP000005205 UP000078492 UP000000311 UP000007755 UP000037069 UP000078541 UP000078540 UP000078542 UP000030742 UP000075809 UP000008237 UP000000304 UP000002282 UP000000803 UP000009192 UP000008792 UP000008711
UP000005204 UP000183832 UP000076502 UP000053825 UP000002358 UP000005203 UP000095301 UP000007062 UP000007266 UP000076407 UP000075883 UP000075903 UP000075886 UP000075880 UP000027135 UP000242457 UP000053105 UP000008820 UP000007819 UP000192223 UP000095300 UP000002320 UP000053097 UP000279307 UP000005205 UP000078492 UP000000311 UP000007755 UP000037069 UP000078541 UP000078540 UP000078542 UP000030742 UP000075809 UP000008237 UP000000304 UP000002282 UP000000803 UP000009192 UP000008792 UP000008711
Pfam
PF07690 MFS_1
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2H1WT49
A0A3S2LLF7
A0A2A4IXD4
A0A194PYJ5
A0A212FAM1
A0A0L7LL21
+ More
A0A212FAL1 A0A194PXP8 A0A194RE89 A0A3S2PBF1 A0A194RDJ0 A0A212FAJ8 A0A212FAK5 A0A194PXM6 A0A194REM1 A0A2A4IY18 A0A3S2LGU6 A0A2W1BZD6 A0A146LMI6 A0A023F3V3 A0A069DVE7 H9JAJ5 A0A2H1W938 A0A1J1HPT6 A0A194QAI0 A0A154PAY0 A0A0L7RGJ1 K7J2K2 A0A088ANF5 A0A1I8M3Q0 Q5TSW5 A0A1B6CY04 A0A2M3Z363 A0A2M4BIV5 A0A2M4BIX1 A0A2M3Z330 A0A1B6LYH9 D6WRR1 A0A2C9H8S5 A0A182MSS2 A0A182VHT3 A0A2M4CUE7 A0A2M4AAV7 A0A2M4CSN5 A0A2M4CUE3 A0A2M4CTY3 A0A182QCE8 A0A182J2E5 A0A212EJ99 A0A212EU49 A0A1Y1KH29 A0A067RJA9 A0A2A3EDD3 A0A0M8ZQJ4 Q16W79 A0A2S2R688 J9KB87 A0A2H8TM94 A0A1W4WW74 A0A1I8PUY1 W8C3P0 A0A1L8DDP9 A0A1Q3FLB8 A0A1Q3FLC7 E9IUT7 B0WAI1 A0A026WMI6 A0A194RDI4 A0A034W601 A0A158NYI1 A0A151ITA2 D2A535 E2A659 F4WT02 A0A0L0C7Q4 A0A195F6C4 A0A195AZB4 A0A195CVZ7 A0A194PXN1 A0A0K8WKB4 U4URR0 A0A2H1WJM3 A0A151WHP4 A0A2S2PEA7 Q6NPA2 A0A194Q3W9 E2BSZ7 A0A2A4IW55 A0A2H1V1J8 A0A0A1XR69 B4R4V6 A0A2H8TQH7 B4PW63 Q9VY22 B4L4F7 A0A1W4XEM7 X1WII3 B4M2P7 A0A1B6HTG8 B3NUZ2
A0A212FAL1 A0A194PXP8 A0A194RE89 A0A3S2PBF1 A0A194RDJ0 A0A212FAJ8 A0A212FAK5 A0A194PXM6 A0A194REM1 A0A2A4IY18 A0A3S2LGU6 A0A2W1BZD6 A0A146LMI6 A0A023F3V3 A0A069DVE7 H9JAJ5 A0A2H1W938 A0A1J1HPT6 A0A194QAI0 A0A154PAY0 A0A0L7RGJ1 K7J2K2 A0A088ANF5 A0A1I8M3Q0 Q5TSW5 A0A1B6CY04 A0A2M3Z363 A0A2M4BIV5 A0A2M4BIX1 A0A2M3Z330 A0A1B6LYH9 D6WRR1 A0A2C9H8S5 A0A182MSS2 A0A182VHT3 A0A2M4CUE7 A0A2M4AAV7 A0A2M4CSN5 A0A2M4CUE3 A0A2M4CTY3 A0A182QCE8 A0A182J2E5 A0A212EJ99 A0A212EU49 A0A1Y1KH29 A0A067RJA9 A0A2A3EDD3 A0A0M8ZQJ4 Q16W79 A0A2S2R688 J9KB87 A0A2H8TM94 A0A1W4WW74 A0A1I8PUY1 W8C3P0 A0A1L8DDP9 A0A1Q3FLB8 A0A1Q3FLC7 E9IUT7 B0WAI1 A0A026WMI6 A0A194RDI4 A0A034W601 A0A158NYI1 A0A151ITA2 D2A535 E2A659 F4WT02 A0A0L0C7Q4 A0A195F6C4 A0A195AZB4 A0A195CVZ7 A0A194PXN1 A0A0K8WKB4 U4URR0 A0A2H1WJM3 A0A151WHP4 A0A2S2PEA7 Q6NPA2 A0A194Q3W9 E2BSZ7 A0A2A4IW55 A0A2H1V1J8 A0A0A1XR69 B4R4V6 A0A2H8TQH7 B4PW63 Q9VY22 B4L4F7 A0A1W4XEM7 X1WII3 B4M2P7 A0A1B6HTG8 B3NUZ2
Ontologies
GO
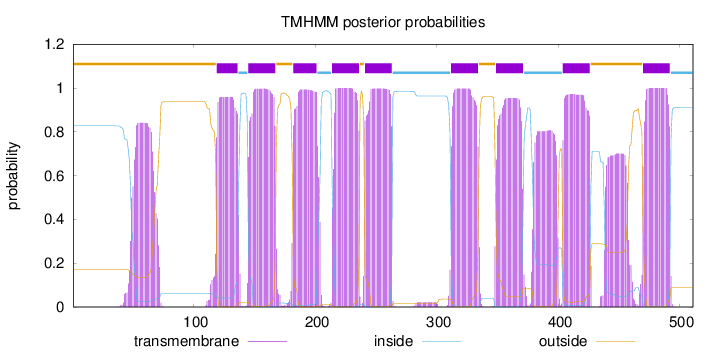
Topology
Length:
511
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
243.30532
Exp number, first 60 AAs:
9.96354
Total prob of N-in:
0.82734
outside
1 - 118
TMhelix
119 - 136
inside
137 - 144
TMhelix
145 - 167
outside
168 - 181
TMhelix
182 - 201
inside
202 - 213
TMhelix
214 - 236
outside
237 - 240
TMhelix
241 - 263
inside
264 - 311
TMhelix
312 - 334
outside
335 - 348
TMhelix
349 - 371
inside
372 - 403
TMhelix
404 - 426
outside
427 - 469
TMhelix
470 - 492
inside
493 - 511
Population Genetic Test Statistics
Pi
19.928066
Theta
25.261825
Tajima's D
-0.560834
CLR
1.015209
CSRT
0.228188590570471
Interpretation
Uncertain
