Gene
KWMTBOMO13370 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010941
Annotation
PREDICTED:_signal_recognition_particle_subunit_SRP72_[Amyelois_transitella]
Full name
Signal recognition particle subunit SRP72
Location in the cell
Cytoplasmic Reliability : 1.645 Nuclear Reliability : 2.163
Sequence
CDS
ATGTCAGCCAACAAGGAGAATAACCTTGTTCAGGCTTATCTTGAATTAAATAAATTTTGCCAGAGCAGTGACTATGAAAGAGCTTTAAAAGCTGCAGGGAAAATACTGCAGATAGCACCAAATGAGCAGAAAGCATTTCATTGCAAAGTGGTATGCTTTCTCCAACTCCACAACTTCAAGGAGGCTCTAGCGACATTAACCAATGCCAAGAACTCAGCGTTAGCAGCAGACCTGCTCTTTGAGAAGGCATACGCTCAATACCGTCTCAATTCACCTAAAGAAGCACTACAAACTGTTGATAGTGCCCCTGAATTGACACCCGCTTTAAAGGAATTAAGGGCTCAAATTTTATACCGCTTAGAGCAATACCAAGACTGCTACAATTTATACCGTGATCTACTGAAGAACACCACTGATGAGTATGAAGATGAGAGGAAAGCCAACATGGCCGCTGTTGTAGCTAACTTGGCAGCACTTAATCCAACGTCAGAGCTGCCGCAGTTTGATGAAAACACATACGAATTGGCATACAACTCGGGCAGCACTTTGGCAATGCGGGGGAAGTATAATGAAGCTTTGTCAGTCTTAAAGAGAGCCGAACAAGCCTGTTCTGAGAGTGTAATAGATGATGGAGGCACTGAGGAAGAGGCTAAAGAAGAAGCTGCCATTATAAGGGTCCAACAAGCTTATTGTCTTCAGCAAATAAACAGGGAGAAGGAAGCGGCTACTATTTACCAGAACATTCTGAAGGACAAACCCAGCGATCAGGCATTAGTCGCGATTGCTAGCAATAATATCATTGTTATAAACAGGGACACGAACGTGTTCGATTCTCGTAAACGCATGAAGGCGGCCACTGCGGATGGACTAGAACACAAACTAAACTCCCGTCAACGAGCTGTCATCACTTATAATCAAGCCATACTCTCTATTTACTCTAATCAACCGGACTTCTGCAAGCAGTGCTGTGTGAAACTGGTCCGTGACTTCGGCGAGGATCGTCGCGCCGCCATTGTGGAGGCGTCCTCGCTCGTCAAGGAAGGGAAGGCATCGCAGGCAGTCAGCTTACTGCTCAAACACGGAGGGACATTGACATTGGCAGCCGTACAGATACTACTCGTTAATGGTGACCGTAAAGCAGCGATCAAGCTGTTACAAGAGAGCGAGTACAAGCATCGTCCCGGAGTCGTGGGAGTGCTGTGCACGCTGCTCTGTGCAGACAACGAGCTGGAGAAGGCCTCGCTGATGTTCGATGAACTATACGAGCACGTCAAGAACGATGCAGAGAGGATGAAGCTGTACCGCGGGGCGTGGCGGGGCGCGGCGCGGTGCCACGGGCGGGCGGGGCGCGCGGGGGCGGCGGCGCGCGCGCACGAGGCGCTGGCGGCGGTGGAGCCGCGGGACGCGCGCAGCCTGGCGCGGCTGGTGAAGGCGCTGGCGGCCGCCGACCCGCCCCGCGCCCGGAGCCTGGCCGAACAGCTGCCGCCCTTGGACCAGCTCGAGACGAAAATCGATATTGACGCCCTGGAATCGTCGAAGTGGATGATGGGAGCTAAAGTCGTCAAGAAGACTGTTCAAAGCAAGCAGGAACAATCCCCCGGAACTCCCGGCTCTGAGTTGGGACAGAAGAAGAAAACGAAACGCAAGCGCAACAGCAAACTCCCGCCGAACGCAGACCTCTCCAAACCTCCCGATCCGGAAAGGTGGTTACCAAAGTACGAGCGAACTGCGTACAGAAAGCGACGAGGCATTAGACGTGACGTCATCAAGGGTAGCCAAGGAATGTCCACCACTGCCACGGATCAATATGACATGAGTAAACAGCAGACCACGCCTACCCCGGCGACCTCTGCCAAGAGTCCCCGGGTCGAAGTCAAGCAGACCGAGAGCGCTTGGCAGCGCAAGCAACAGCAGAAGAAGAAAGGCAAGGGACGTAAATGGTAG
Protein
MSANKENNLVQAYLELNKFCQSSDYERALKAAGKILQIAPNEQKAFHCKVVCFLQLHNFKEALATLTNAKNSALAADLLFEKAYAQYRLNSPKEALQTVDSAPELTPALKELRAQILYRLEQYQDCYNLYRDLLKNTTDEYEDERKANMAAVVANLAALNPTSELPQFDENTYELAYNSGSTLAMRGKYNEALSVLKRAEQACSESVIDDGGTEEEAKEEAAIIRVQQAYCLQQINREKEAATIYQNILKDKPSDQALVAIASNNIIVINRDTNVFDSRKRMKAATADGLEHKLNSRQRAVITYNQAILSIYSNQPDFCKQCCVKLVRDFGEDRRAAIVEASSLVKEGKASQAVSLLLKHGGTLTLAAVQILLVNGDRKAAIKLLQESEYKHRPGVVGVLCTLLCADNELEKASLMFDELYEHVKNDAERMKLYRGAWRGAARCHGRAGRAGAAARAHEALAAVEPRDARSLARLVKALAAADPPRARSLAEQLPPLDQLETKIDIDALESSKWMMGAKVVKKTVQSKQEQSPGTPGSELGQKKKTKRKRNSKLPPNADLSKPPDPERWLPKYERTAYRKRRGIRRDVIKGSQGMSTTATDQYDMSKQQTTPTPATSAKSPRVEVKQTESAWQRKQQQKKKGKGRKW
Summary
Description
Signal-recognition-particle assembly has a crucial role in targeting secretory proteins to the rough endoplasmic reticulum membrane.
Similarity
Belongs to the SRP72 family.
Uniprot
A0A2H1V6P4
S4P286
A0A212FAZ2
A0A194PZD6
A0A194REL1
A0A067RSF1
+ More
A0A2J7PZF5 A0A0C9R6H6 A0A1B6F764 A0A0K8TLY5 E0VYM3 A0A0L7QN09 A0A0J7KZ88 E2AD30 A0A1Q3F9F8 E2BIP9 B0XFF2 A0A1L8DW84 A0A182G6Q7 A0A195FKD6 A0A1I8MHA3 T1PET2 A0A0M9A046 A0A151IG22 A0A1B0CFH1 A0A151IXR6 Q17FK4 A0A182G1E5 A0A1I8NU27 A0A232F8D1 D3TM83 U5EXU8 E9IAT5 A0A2A3EFH3 A0A1A9VB90 A0A1A9ZHL9 A0A1B0BNL0 K7IR24 A0A1A9XGA0 A0A154PKL0 A0A026VVC8 A0A034WNJ2 A0A1B0G7Y6 A0A0A1WGI1 A0A0K8W570 W8BIH7 A0A195BAN3 F4WRA0 A0A088A4L7 A0A1B0DK34 A0A0P4WFU4 T1IQJ0 A0A131XGM6 A0A1E1XRG6 L7MA07 A0A023GNZ0 A0A3M7P871 A0A3Q3IWM8 G3MFM4 I3IYX9 A0A3Q2X3D2 A0A3Q1HXB1 A0A218UNK1 R0JCR9 U3JW70 A0A3P9B384 A0A3P8PXC2 A0A3B4EWF4 A0A1U7RR68 U3IMM8 A0A1A8DJV1 G3PWX0 A0A1A8AH52 A0A1A8HNT3 A0A1V4JSV7 A0A1A8RS77 A0A091E493 A0A3B4YEZ5 Q5ZKZ5 A0A2I4CG33 A0A1V4JSU5 A0A0Q3PKI8 A0A3B5ABY4 A0A087XCI9 A0A3B3UTR6 A0A1U7SPL9 A0A286X7B1 G3VBY3 A0A2I3S3Y1 A0A384CY45 A0A3Q7XPF4 A0A1A7YYU5 A0A383YRC2 K7DN16 A0A2R9CBA5 K7CCE7 V9HWK0
A0A2J7PZF5 A0A0C9R6H6 A0A1B6F764 A0A0K8TLY5 E0VYM3 A0A0L7QN09 A0A0J7KZ88 E2AD30 A0A1Q3F9F8 E2BIP9 B0XFF2 A0A1L8DW84 A0A182G6Q7 A0A195FKD6 A0A1I8MHA3 T1PET2 A0A0M9A046 A0A151IG22 A0A1B0CFH1 A0A151IXR6 Q17FK4 A0A182G1E5 A0A1I8NU27 A0A232F8D1 D3TM83 U5EXU8 E9IAT5 A0A2A3EFH3 A0A1A9VB90 A0A1A9ZHL9 A0A1B0BNL0 K7IR24 A0A1A9XGA0 A0A154PKL0 A0A026VVC8 A0A034WNJ2 A0A1B0G7Y6 A0A0A1WGI1 A0A0K8W570 W8BIH7 A0A195BAN3 F4WRA0 A0A088A4L7 A0A1B0DK34 A0A0P4WFU4 T1IQJ0 A0A131XGM6 A0A1E1XRG6 L7MA07 A0A023GNZ0 A0A3M7P871 A0A3Q3IWM8 G3MFM4 I3IYX9 A0A3Q2X3D2 A0A3Q1HXB1 A0A218UNK1 R0JCR9 U3JW70 A0A3P9B384 A0A3P8PXC2 A0A3B4EWF4 A0A1U7RR68 U3IMM8 A0A1A8DJV1 G3PWX0 A0A1A8AH52 A0A1A8HNT3 A0A1V4JSV7 A0A1A8RS77 A0A091E493 A0A3B4YEZ5 Q5ZKZ5 A0A2I4CG33 A0A1V4JSU5 A0A0Q3PKI8 A0A3B5ABY4 A0A087XCI9 A0A3B3UTR6 A0A1U7SPL9 A0A286X7B1 G3VBY3 A0A2I3S3Y1 A0A384CY45 A0A3Q7XPF4 A0A1A7YYU5 A0A383YRC2 K7DN16 A0A2R9CBA5 K7CCE7 V9HWK0
Pubmed
23622113
22118469
26354079
24845553
26369729
20566863
+ More
20798317 26483478 25315136 17510324 28648823 20353571 21282665 20075255 24508170 30249741 25348373 25830018 24495485 21719571 28049606 29209593 25576852 30375419 22216098 25186727 23749191 15592404 15642098 21993624 21709235 16136131 24813606 22722832 11181995
20798317 26483478 25315136 17510324 28648823 20353571 21282665 20075255 24508170 30249741 25348373 25830018 24495485 21719571 28049606 29209593 25576852 30375419 22216098 25186727 23749191 15592404 15642098 21993624 21709235 16136131 24813606 22722832 11181995
EMBL
ODYU01000963
SOQ36513.1
GAIX01012145
JAA80415.1
AGBW02009396
OWR50915.1
+ More
KQ459586 KPI98104.1 KQ460323 KPJ15899.1 KK852446 KDR23740.1 NEVH01020335 PNF21717.1 GBYB01011934 JAG81701.1 GECZ01023661 JAS46108.1 GDAI01002467 JAI15136.1 DS235845 EEB18479.1 KQ414885 KOC59861.1 LBMM01001886 KMQ95608.1 GL438658 EFN68666.1 GFDL01010830 JAV24215.1 GL448520 EFN84443.1 DS232921 EDS26725.1 GFDF01003579 JAV10505.1 JXUM01148643 JXUM01148644 KQ570607 KXJ68334.1 KQ981512 KYN40821.1 KA647204 AFP61833.1 KQ435803 KOX73099.1 KQ977739 KYN00219.1 AJWK01010103 AJWK01010104 AJWK01010105 AJWK01010106 KQ980795 KYN13028.1 CH477270 EAT45339.1 JXUM01136598 KQ568453 KXJ68994.1 NNAY01000669 OXU27091.1 EZ422535 ADD18811.1 GANO01000742 JAB59129.1 GL762061 EFZ22318.1 KZ288266 PBC30234.1 JXJN01017452 KQ434948 KZC12385.1 KK107796 QOIP01000013 EZA47743.1 RLU15374.1 GAKP01003611 GAKP01003609 JAC55343.1 CCAG010010579 GBXI01016163 JAC98128.1 GDHF01005976 JAI46338.1 GAMC01009737 JAB96818.1 KQ976532 KYM81598.1 GL888284 EGI63317.1 AJVK01035231 GDRN01060279 JAI65393.1 AFFK01018331 GEFH01003333 JAP65248.1 GFAA01001524 JAU01911.1 GACK01005085 JAA59949.1 GBBM01000613 JAC34805.1 REGN01012711 RMZ94947.1 JO840675 AEO32292.1 AERX01009965 MUZQ01000223 OWK54942.1 KB744596 EOA94766.1 AGTO01021376 ADON01132353 HADZ01016045 HAEA01005298 SBQ33778.1 HADY01015070 SBP53555.1 HAED01000226 SBQ86071.1 LSYS01006437 OPJ75278.1 HAEI01009828 HAEH01021791 SBS08886.1 KN120907 KFO37385.1 AADN05000017 AJ719939 CAG31598.1 OPJ75279.1 LMAW01002336 KQK81523.1 AYCK01014770 AYCK01014771 AAKN02028318 AEFK01224946 AEFK01224947 AACZ04033994 HADX01013167 SBP35399.1 GABE01002719 JAA42020.1 AJFE02017999 AJFE02018000 AJFE02018001 GABF01000270 NBAG03000215 JAA21875.1 PNI82513.1 EU794663 CH471057 ACJ13717.1 EAX05496.1
KQ459586 KPI98104.1 KQ460323 KPJ15899.1 KK852446 KDR23740.1 NEVH01020335 PNF21717.1 GBYB01011934 JAG81701.1 GECZ01023661 JAS46108.1 GDAI01002467 JAI15136.1 DS235845 EEB18479.1 KQ414885 KOC59861.1 LBMM01001886 KMQ95608.1 GL438658 EFN68666.1 GFDL01010830 JAV24215.1 GL448520 EFN84443.1 DS232921 EDS26725.1 GFDF01003579 JAV10505.1 JXUM01148643 JXUM01148644 KQ570607 KXJ68334.1 KQ981512 KYN40821.1 KA647204 AFP61833.1 KQ435803 KOX73099.1 KQ977739 KYN00219.1 AJWK01010103 AJWK01010104 AJWK01010105 AJWK01010106 KQ980795 KYN13028.1 CH477270 EAT45339.1 JXUM01136598 KQ568453 KXJ68994.1 NNAY01000669 OXU27091.1 EZ422535 ADD18811.1 GANO01000742 JAB59129.1 GL762061 EFZ22318.1 KZ288266 PBC30234.1 JXJN01017452 KQ434948 KZC12385.1 KK107796 QOIP01000013 EZA47743.1 RLU15374.1 GAKP01003611 GAKP01003609 JAC55343.1 CCAG010010579 GBXI01016163 JAC98128.1 GDHF01005976 JAI46338.1 GAMC01009737 JAB96818.1 KQ976532 KYM81598.1 GL888284 EGI63317.1 AJVK01035231 GDRN01060279 JAI65393.1 AFFK01018331 GEFH01003333 JAP65248.1 GFAA01001524 JAU01911.1 GACK01005085 JAA59949.1 GBBM01000613 JAC34805.1 REGN01012711 RMZ94947.1 JO840675 AEO32292.1 AERX01009965 MUZQ01000223 OWK54942.1 KB744596 EOA94766.1 AGTO01021376 ADON01132353 HADZ01016045 HAEA01005298 SBQ33778.1 HADY01015070 SBP53555.1 HAED01000226 SBQ86071.1 LSYS01006437 OPJ75278.1 HAEI01009828 HAEH01021791 SBS08886.1 KN120907 KFO37385.1 AADN05000017 AJ719939 CAG31598.1 OPJ75279.1 LMAW01002336 KQK81523.1 AYCK01014770 AYCK01014771 AAKN02028318 AEFK01224946 AEFK01224947 AACZ04033994 HADX01013167 SBP35399.1 GABE01002719 JAA42020.1 AJFE02017999 AJFE02018000 AJFE02018001 GABF01000270 NBAG03000215 JAA21875.1 PNI82513.1 EU794663 CH471057 ACJ13717.1 EAX05496.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000027135
UP000235965
UP000009046
+ More
UP000053825 UP000036403 UP000000311 UP000008237 UP000002320 UP000069940 UP000249989 UP000078541 UP000095301 UP000053105 UP000078542 UP000092461 UP000078492 UP000008820 UP000095300 UP000215335 UP000242457 UP000078200 UP000092445 UP000092460 UP000002358 UP000092443 UP000076502 UP000053097 UP000279307 UP000092444 UP000078540 UP000007755 UP000005203 UP000092462 UP000276133 UP000261600 UP000005207 UP000264840 UP000265040 UP000197619 UP000016665 UP000265160 UP000265100 UP000261460 UP000189705 UP000016666 UP000007635 UP000190648 UP000028990 UP000261360 UP000000539 UP000192220 UP000051836 UP000261400 UP000028760 UP000261500 UP000189704 UP000005447 UP000007648 UP000002277 UP000261680 UP000291021 UP000286642 UP000261681 UP000240080
UP000053825 UP000036403 UP000000311 UP000008237 UP000002320 UP000069940 UP000249989 UP000078541 UP000095301 UP000053105 UP000078542 UP000092461 UP000078492 UP000008820 UP000095300 UP000215335 UP000242457 UP000078200 UP000092445 UP000092460 UP000002358 UP000092443 UP000076502 UP000053097 UP000279307 UP000092444 UP000078540 UP000007755 UP000005203 UP000092462 UP000276133 UP000261600 UP000005207 UP000264840 UP000265040 UP000197619 UP000016665 UP000265160 UP000265100 UP000261460 UP000189705 UP000016666 UP000007635 UP000190648 UP000028990 UP000261360 UP000000539 UP000192220 UP000051836 UP000261400 UP000028760 UP000261500 UP000189704 UP000005447 UP000007648 UP000002277 UP000261680 UP000291021 UP000286642 UP000261681 UP000240080
PRIDE
Pfam
Interpro
IPR019734
TPR_repeat
+ More
IPR011990 TPR-like_helical_dom_sf
IPR026270 SRP72
IPR031545 SRP_TPR-like
IPR013699 Signal_recog_part_SRP72_RNA-bd
IPR013026 TPR-contain_dom
IPR018704 TPR_21
IPR015374 ChAPs
IPR012934 Znf_AD
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR036728 PBP_GOBP_sf
IPR006170 PBP/GOBP
IPR002885 Pentatricopeptide_repeat
IPR027417 P-loop_NTPase
IPR006689 Small_GTPase_ARF/SAR
IPR011990 TPR-like_helical_dom_sf
IPR026270 SRP72
IPR031545 SRP_TPR-like
IPR013699 Signal_recog_part_SRP72_RNA-bd
IPR013026 TPR-contain_dom
IPR018704 TPR_21
IPR015374 ChAPs
IPR012934 Znf_AD
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR036728 PBP_GOBP_sf
IPR006170 PBP/GOBP
IPR002885 Pentatricopeptide_repeat
IPR027417 P-loop_NTPase
IPR006689 Small_GTPase_ARF/SAR
Gene 3D
ProteinModelPortal
A0A2H1V6P4
S4P286
A0A212FAZ2
A0A194PZD6
A0A194REL1
A0A067RSF1
+ More
A0A2J7PZF5 A0A0C9R6H6 A0A1B6F764 A0A0K8TLY5 E0VYM3 A0A0L7QN09 A0A0J7KZ88 E2AD30 A0A1Q3F9F8 E2BIP9 B0XFF2 A0A1L8DW84 A0A182G6Q7 A0A195FKD6 A0A1I8MHA3 T1PET2 A0A0M9A046 A0A151IG22 A0A1B0CFH1 A0A151IXR6 Q17FK4 A0A182G1E5 A0A1I8NU27 A0A232F8D1 D3TM83 U5EXU8 E9IAT5 A0A2A3EFH3 A0A1A9VB90 A0A1A9ZHL9 A0A1B0BNL0 K7IR24 A0A1A9XGA0 A0A154PKL0 A0A026VVC8 A0A034WNJ2 A0A1B0G7Y6 A0A0A1WGI1 A0A0K8W570 W8BIH7 A0A195BAN3 F4WRA0 A0A088A4L7 A0A1B0DK34 A0A0P4WFU4 T1IQJ0 A0A131XGM6 A0A1E1XRG6 L7MA07 A0A023GNZ0 A0A3M7P871 A0A3Q3IWM8 G3MFM4 I3IYX9 A0A3Q2X3D2 A0A3Q1HXB1 A0A218UNK1 R0JCR9 U3JW70 A0A3P9B384 A0A3P8PXC2 A0A3B4EWF4 A0A1U7RR68 U3IMM8 A0A1A8DJV1 G3PWX0 A0A1A8AH52 A0A1A8HNT3 A0A1V4JSV7 A0A1A8RS77 A0A091E493 A0A3B4YEZ5 Q5ZKZ5 A0A2I4CG33 A0A1V4JSU5 A0A0Q3PKI8 A0A3B5ABY4 A0A087XCI9 A0A3B3UTR6 A0A1U7SPL9 A0A286X7B1 G3VBY3 A0A2I3S3Y1 A0A384CY45 A0A3Q7XPF4 A0A1A7YYU5 A0A383YRC2 K7DN16 A0A2R9CBA5 K7CCE7 V9HWK0
A0A2J7PZF5 A0A0C9R6H6 A0A1B6F764 A0A0K8TLY5 E0VYM3 A0A0L7QN09 A0A0J7KZ88 E2AD30 A0A1Q3F9F8 E2BIP9 B0XFF2 A0A1L8DW84 A0A182G6Q7 A0A195FKD6 A0A1I8MHA3 T1PET2 A0A0M9A046 A0A151IG22 A0A1B0CFH1 A0A151IXR6 Q17FK4 A0A182G1E5 A0A1I8NU27 A0A232F8D1 D3TM83 U5EXU8 E9IAT5 A0A2A3EFH3 A0A1A9VB90 A0A1A9ZHL9 A0A1B0BNL0 K7IR24 A0A1A9XGA0 A0A154PKL0 A0A026VVC8 A0A034WNJ2 A0A1B0G7Y6 A0A0A1WGI1 A0A0K8W570 W8BIH7 A0A195BAN3 F4WRA0 A0A088A4L7 A0A1B0DK34 A0A0P4WFU4 T1IQJ0 A0A131XGM6 A0A1E1XRG6 L7MA07 A0A023GNZ0 A0A3M7P871 A0A3Q3IWM8 G3MFM4 I3IYX9 A0A3Q2X3D2 A0A3Q1HXB1 A0A218UNK1 R0JCR9 U3JW70 A0A3P9B384 A0A3P8PXC2 A0A3B4EWF4 A0A1U7RR68 U3IMM8 A0A1A8DJV1 G3PWX0 A0A1A8AH52 A0A1A8HNT3 A0A1V4JSV7 A0A1A8RS77 A0A091E493 A0A3B4YEZ5 Q5ZKZ5 A0A2I4CG33 A0A1V4JSU5 A0A0Q3PKI8 A0A3B5ABY4 A0A087XCI9 A0A3B3UTR6 A0A1U7SPL9 A0A286X7B1 G3VBY3 A0A2I3S3Y1 A0A384CY45 A0A3Q7XPF4 A0A1A7YYU5 A0A383YRC2 K7DN16 A0A2R9CBA5 K7CCE7 V9HWK0
PDB
6FRK
E-value=3.49645e-77,
Score=736
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
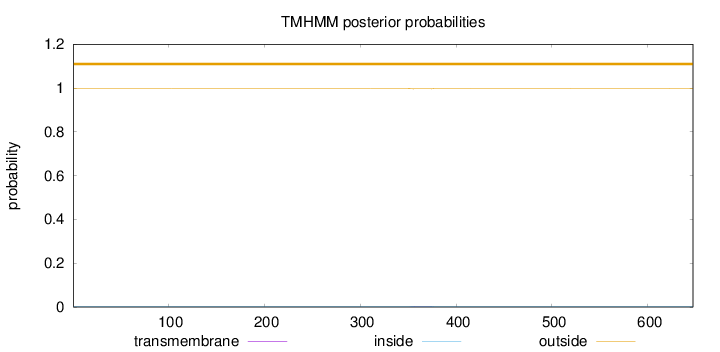
Length:
647
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05124
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.00237
outside
1 - 647
Population Genetic Test Statistics
Pi
26.309953
Theta
23.595731
Tajima's D
0.155186
CLR
0.36164
CSRT
0.419479026048698
Interpretation
Uncertain
