Gene
KWMTBOMO13365 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010943
Annotation
ADP-ribosylation_factor-like_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.088 Nuclear Reliability : 1.172
Sequence
CDS
ATGCTTGCTTTAATCAATCGCATCTTAGATTGGATTAAAAGCTTGTTTTGGAAGGAGGAAATGGAGCTTACACTGGTGGGCTTGCAGTATTCGGGCAAAACAACTTTTGTTAACGTCATTGCTTCAGGCCAGTTTAGTGAAGACATGATTCCAACAGTCGGTTTCAACATGCGCAAAGTAACCAAAGGAAACGTAACAATCAAGGTATGGGATATTGGTGGCCAACCAAGATTCCGTTCAATGTGGGAGCGTTATTGCAGAGGGGTCAATGCTATTGTGTACATGGTGGACGCGGCCGACCCTGATAAGATCGAGGCGTCGCGTAACGAGCTTCACAGCCTGCTGGAGAAGCAGCAGCTCACCGGCATCCCCGTGCTGGTGCTCGGCAACAAGCGCGATCTGCCACAAGCTCTCGACGAACATGGACTCATTGAAAGAATGAACCTGTCAGCGATCCAGGACCGCGAGATTTGCTGTTATTCCATATCGTGCAAGGAGAAAGACAACATCGACATAACTCTCCAGTGGCTGATCGCGCACAGTAAGTCTGGTAGCGCGCATTAA
Protein
MLALINRILDWIKSLFWKEEMELTLVGLQYSGKTTFVNVIASGQFSEDMIPTVGFNMRKVTKGNVTIKVWDIGGQPRFRSMWERYCRGVNAIVYMVDAADPDKIEASRNELHSLLEKQQLTGIPVLVLGNKRDLPQALDEHGLIERMNLSAIQDREICCYSISCKEKDNIDITLQWLIAHSKSGSAH
Summary
Similarity
Belongs to the small GTPase superfamily. Arf family.
Uniprot
Q2F632
A0A2A4IWF6
A0A194PZE1
A0A1E1W6B7
A0A2H1W5K1
A0A194REK4
+ More
A0A212ESR9 A0A067RKX8 D6WZI7 A0A154PG00 A0A3S2NJN3 A0A2P2I0D2 V5H2S4 A0A0P4WH28 N6TVS1 A0A1Y1KEG3 A0A195DDT3 E9IXZ0 E2B6I2 A0A158P3L8 E1ZUV5 A0A151IGW4 A0A195FUB0 A0A026WY30 A0A232F8S4 K7J7Y2 A0A2A3EKQ5 V9IID2 A0A088AIA6 A0A151WFF0 A0A1W4XHD2 A0A0C9PZW3 A0A310SHX4 A0A0M9A9G4 A0A2M4ALX5 A0A0L7RDL5 A0A1I8NNG4 A0A0K8TNG8 A0A0L0CJD8 T1E326 A0A2M3ZIS5 A0A2M4C110 A0A182FPD4 A0A1Q3EU66 A0A034W4V4 A0A0C9R9J3 W5JLE3 A0A1I8N822 U5EZE1 A0A023EIL1 A0A1L8DVV3 A0A1B0CNK8 A0A0A1WNA3 W8BI58 E9GY65 A0A0P6F0K2 A0A182P004 B0X8S5 B4M158 B4K966 A0A0M5J1E2 A0A1A9X5Y4 A0A1A9V5I4 A0A1B0ASN6 A0A1A9Z747 D3TQV5 A0A3B0KQN3 Q299A4 B4G4Y2 B3LZH3 W5USM2 A0A0K8WFW5 Q172Z9 A0A182Y5F1 A0A182M7N7 A0A182Q111 A0A182JXU4 A0A182U5U0 A0A182W1B8 A0A182XG61 Q7PQM1 A0A182RV76 A0A182IDA6 A0A084VLD7 A0A182J175 A0A3Q2PX03 B4NJ88 A0A1J1HJ99 A0A3B3CVH6 A0A3Q3GWC8 A0A1A7X530 A0A1B6C5J4 A0A1A8PDA8 A0A1A8JPM4 A0A1A8E856 A0A1A8HAX4 A0A1A8VH75 A0A1W4W2Y2 B4QYQ2 B4PSL3
A0A212ESR9 A0A067RKX8 D6WZI7 A0A154PG00 A0A3S2NJN3 A0A2P2I0D2 V5H2S4 A0A0P4WH28 N6TVS1 A0A1Y1KEG3 A0A195DDT3 E9IXZ0 E2B6I2 A0A158P3L8 E1ZUV5 A0A151IGW4 A0A195FUB0 A0A026WY30 A0A232F8S4 K7J7Y2 A0A2A3EKQ5 V9IID2 A0A088AIA6 A0A151WFF0 A0A1W4XHD2 A0A0C9PZW3 A0A310SHX4 A0A0M9A9G4 A0A2M4ALX5 A0A0L7RDL5 A0A1I8NNG4 A0A0K8TNG8 A0A0L0CJD8 T1E326 A0A2M3ZIS5 A0A2M4C110 A0A182FPD4 A0A1Q3EU66 A0A034W4V4 A0A0C9R9J3 W5JLE3 A0A1I8N822 U5EZE1 A0A023EIL1 A0A1L8DVV3 A0A1B0CNK8 A0A0A1WNA3 W8BI58 E9GY65 A0A0P6F0K2 A0A182P004 B0X8S5 B4M158 B4K966 A0A0M5J1E2 A0A1A9X5Y4 A0A1A9V5I4 A0A1B0ASN6 A0A1A9Z747 D3TQV5 A0A3B0KQN3 Q299A4 B4G4Y2 B3LZH3 W5USM2 A0A0K8WFW5 Q172Z9 A0A182Y5F1 A0A182M7N7 A0A182Q111 A0A182JXU4 A0A182U5U0 A0A182W1B8 A0A182XG61 Q7PQM1 A0A182RV76 A0A182IDA6 A0A084VLD7 A0A182J175 A0A3Q2PX03 B4NJ88 A0A1J1HJ99 A0A3B3CVH6 A0A3Q3GWC8 A0A1A7X530 A0A1B6C5J4 A0A1A8PDA8 A0A1A8JPM4 A0A1A8E856 A0A1A8HAX4 A0A1A8VH75 A0A1W4W2Y2 B4QYQ2 B4PSL3
Pubmed
19121390
26354079
22118469
24845553
18362917
19820115
+ More
23537049 28004739 21282665 20798317 21347285 24508170 30249741 28648823 20075255 26369729 26108605 24330624 25348373 20920257 23761445 25315136 24945155 26483478 25830018 24495485 21292972 17994087 20353571 15632085 23127152 17510324 25244985 12364791 14747013 17210077 24438588 29451363 17550304
23537049 28004739 21282665 20798317 21347285 24508170 30249741 28648823 20075255 26369729 26108605 24330624 25348373 20920257 23761445 25315136 24945155 26483478 25830018 24495485 21292972 17994087 20353571 15632085 23127152 17510324 25244985 12364791 14747013 17210077 24438588 29451363 17550304
EMBL
BABH01030523
DQ311240
ABD36185.1
NWSH01005313
PCG64297.1
KQ459586
+ More
KPI98109.1 GDQN01008522 GDQN01002542 JAT82532.1 JAT88512.1 ODYU01006443 SOQ48293.1 KQ460323 KPJ15894.1 AGBW02012724 OWR44538.1 KK852616 KDR20181.1 KQ971372 EFA10434.1 KQ434896 KZC10771.1 RSAL01000068 RVE49277.1 IACF01001804 LAB67483.1 GALX01001328 JAB67138.1 GDRN01062034 JAI65124.1 APGK01059072 KB741292 KB632349 ENN70397.1 ERL93215.1 GEZM01087375 JAV58731.1 KQ980989 KYN10584.1 GL766836 EFZ14564.1 GL445973 EFN88726.1 ADTU01001333 GL434297 EFN75047.1 KQ977726 KYN00313.1 KQ981264 KYN44011.1 KK107069 QOIP01000002 EZA60728.1 RLU25981.1 NNAY01000641 OXU27241.1 KZ288222 PBC32074.1 JR046792 AEY60251.1 KQ983219 KYQ46554.1 GBYB01007098 GBYB01008880 GBYB01008882 JAG76865.1 JAG78647.1 JAG78649.1 KQ768821 OAD53052.1 KQ435720 KOX78663.1 GGFK01008411 MBW41732.1 KQ414614 KOC68909.1 GDAI01001694 JAI15909.1 JRES01000409 KNC31594.1 GALA01000755 JAA94097.1 GGFM01007661 MBW28412.1 GGFJ01009720 MBW58861.1 GFDL01016198 JAV18847.1 GAKP01010164 JAC48788.1 GBYB01009592 JAG79359.1 ADMH02000740 ETN65197.1 GANO01000962 JAB58909.1 JXUM01026644 JXUM01082715 JXUM01082716 GAPW01004807 KQ563332 KQ560763 JAC08791.1 KXJ74034.1 KXJ80987.1 GFDF01003524 JAV10560.1 AJWK01020464 GBXI01013753 JAD00539.1 GAMC01009917 GAMC01009916 JAB96638.1 GL732574 EFX75547.1 GDIP01233504 GDIQ01054736 LRGB01000872 JAI89897.1 JAN40001.1 KZS15391.1 DS232500 EDS42667.1 CH940650 EDW67469.2 CH933806 EDW14479.1 CP012526 ALC45624.1 JXJN01002887 CCAG010018315 EZ423807 ADD20083.1 OUUW01000013 SPP88166.1 CM000070 EAL27799.1 CH479179 EDW24648.1 CH902617 EDV44152.1 JT418140 AHH42633.1 GDHF01002251 JAI50063.1 CH477428 EAT41129.1 AXCM01002342 AXCN02001573 AAAB01008880 EAA08617.4 APCN01005087 ATLV01014478 KE524973 KFB38781.1 CH964272 EDW84919.1 CVRI01000006 CRK88093.1 HADW01011817 HADX01004975 SBP13217.1 GEDC01028532 JAS08766.1 HAEF01007021 HAEG01007431 SBR79261.1 HAED01011797 HAEE01001996 SBR22016.1 HADZ01004702 HAEA01013892 SBQ42372.1 HAEB01008286 HAEC01013099 SBQ81316.1 HADY01002569 HAEJ01018071 SBS58528.1 CM000364 EDX13810.1 CM000160 EDW96473.1
KPI98109.1 GDQN01008522 GDQN01002542 JAT82532.1 JAT88512.1 ODYU01006443 SOQ48293.1 KQ460323 KPJ15894.1 AGBW02012724 OWR44538.1 KK852616 KDR20181.1 KQ971372 EFA10434.1 KQ434896 KZC10771.1 RSAL01000068 RVE49277.1 IACF01001804 LAB67483.1 GALX01001328 JAB67138.1 GDRN01062034 JAI65124.1 APGK01059072 KB741292 KB632349 ENN70397.1 ERL93215.1 GEZM01087375 JAV58731.1 KQ980989 KYN10584.1 GL766836 EFZ14564.1 GL445973 EFN88726.1 ADTU01001333 GL434297 EFN75047.1 KQ977726 KYN00313.1 KQ981264 KYN44011.1 KK107069 QOIP01000002 EZA60728.1 RLU25981.1 NNAY01000641 OXU27241.1 KZ288222 PBC32074.1 JR046792 AEY60251.1 KQ983219 KYQ46554.1 GBYB01007098 GBYB01008880 GBYB01008882 JAG76865.1 JAG78647.1 JAG78649.1 KQ768821 OAD53052.1 KQ435720 KOX78663.1 GGFK01008411 MBW41732.1 KQ414614 KOC68909.1 GDAI01001694 JAI15909.1 JRES01000409 KNC31594.1 GALA01000755 JAA94097.1 GGFM01007661 MBW28412.1 GGFJ01009720 MBW58861.1 GFDL01016198 JAV18847.1 GAKP01010164 JAC48788.1 GBYB01009592 JAG79359.1 ADMH02000740 ETN65197.1 GANO01000962 JAB58909.1 JXUM01026644 JXUM01082715 JXUM01082716 GAPW01004807 KQ563332 KQ560763 JAC08791.1 KXJ74034.1 KXJ80987.1 GFDF01003524 JAV10560.1 AJWK01020464 GBXI01013753 JAD00539.1 GAMC01009917 GAMC01009916 JAB96638.1 GL732574 EFX75547.1 GDIP01233504 GDIQ01054736 LRGB01000872 JAI89897.1 JAN40001.1 KZS15391.1 DS232500 EDS42667.1 CH940650 EDW67469.2 CH933806 EDW14479.1 CP012526 ALC45624.1 JXJN01002887 CCAG010018315 EZ423807 ADD20083.1 OUUW01000013 SPP88166.1 CM000070 EAL27799.1 CH479179 EDW24648.1 CH902617 EDV44152.1 JT418140 AHH42633.1 GDHF01002251 JAI50063.1 CH477428 EAT41129.1 AXCM01002342 AXCN02001573 AAAB01008880 EAA08617.4 APCN01005087 ATLV01014478 KE524973 KFB38781.1 CH964272 EDW84919.1 CVRI01000006 CRK88093.1 HADW01011817 HADX01004975 SBP13217.1 GEDC01028532 JAS08766.1 HAEF01007021 HAEG01007431 SBR79261.1 HAED01011797 HAEE01001996 SBR22016.1 HADZ01004702 HAEA01013892 SBQ42372.1 HAEB01008286 HAEC01013099 SBQ81316.1 HADY01002569 HAEJ01018071 SBS58528.1 CM000364 EDX13810.1 CM000160 EDW96473.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000027135
+ More
UP000007266 UP000076502 UP000283053 UP000019118 UP000030742 UP000078492 UP000008237 UP000005205 UP000000311 UP000078542 UP000078541 UP000053097 UP000279307 UP000215335 UP000002358 UP000242457 UP000005203 UP000075809 UP000192223 UP000053105 UP000053825 UP000095300 UP000037069 UP000069272 UP000000673 UP000095301 UP000069940 UP000249989 UP000092461 UP000000305 UP000076858 UP000075885 UP000002320 UP000008792 UP000009192 UP000092553 UP000092443 UP000078200 UP000092460 UP000092445 UP000092444 UP000268350 UP000001819 UP000008744 UP000007801 UP000221080 UP000008820 UP000076408 UP000075883 UP000075886 UP000075881 UP000075902 UP000075920 UP000076407 UP000007062 UP000075900 UP000075840 UP000030765 UP000075880 UP000265000 UP000007798 UP000183832 UP000261560 UP000264800 UP000192221 UP000000304 UP000002282
UP000007266 UP000076502 UP000283053 UP000019118 UP000030742 UP000078492 UP000008237 UP000005205 UP000000311 UP000078542 UP000078541 UP000053097 UP000279307 UP000215335 UP000002358 UP000242457 UP000005203 UP000075809 UP000192223 UP000053105 UP000053825 UP000095300 UP000037069 UP000069272 UP000000673 UP000095301 UP000069940 UP000249989 UP000092461 UP000000305 UP000076858 UP000075885 UP000002320 UP000008792 UP000009192 UP000092553 UP000092443 UP000078200 UP000092460 UP000092445 UP000092444 UP000268350 UP000001819 UP000008744 UP000007801 UP000221080 UP000008820 UP000076408 UP000075883 UP000075886 UP000075881 UP000075902 UP000075920 UP000076407 UP000007062 UP000075900 UP000075840 UP000030765 UP000075880 UP000265000 UP000007798 UP000183832 UP000261560 UP000264800 UP000192221 UP000000304 UP000002282
PRIDE
Pfam
PF00025 Arf
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
Q2F632
A0A2A4IWF6
A0A194PZE1
A0A1E1W6B7
A0A2H1W5K1
A0A194REK4
+ More
A0A212ESR9 A0A067RKX8 D6WZI7 A0A154PG00 A0A3S2NJN3 A0A2P2I0D2 V5H2S4 A0A0P4WH28 N6TVS1 A0A1Y1KEG3 A0A195DDT3 E9IXZ0 E2B6I2 A0A158P3L8 E1ZUV5 A0A151IGW4 A0A195FUB0 A0A026WY30 A0A232F8S4 K7J7Y2 A0A2A3EKQ5 V9IID2 A0A088AIA6 A0A151WFF0 A0A1W4XHD2 A0A0C9PZW3 A0A310SHX4 A0A0M9A9G4 A0A2M4ALX5 A0A0L7RDL5 A0A1I8NNG4 A0A0K8TNG8 A0A0L0CJD8 T1E326 A0A2M3ZIS5 A0A2M4C110 A0A182FPD4 A0A1Q3EU66 A0A034W4V4 A0A0C9R9J3 W5JLE3 A0A1I8N822 U5EZE1 A0A023EIL1 A0A1L8DVV3 A0A1B0CNK8 A0A0A1WNA3 W8BI58 E9GY65 A0A0P6F0K2 A0A182P004 B0X8S5 B4M158 B4K966 A0A0M5J1E2 A0A1A9X5Y4 A0A1A9V5I4 A0A1B0ASN6 A0A1A9Z747 D3TQV5 A0A3B0KQN3 Q299A4 B4G4Y2 B3LZH3 W5USM2 A0A0K8WFW5 Q172Z9 A0A182Y5F1 A0A182M7N7 A0A182Q111 A0A182JXU4 A0A182U5U0 A0A182W1B8 A0A182XG61 Q7PQM1 A0A182RV76 A0A182IDA6 A0A084VLD7 A0A182J175 A0A3Q2PX03 B4NJ88 A0A1J1HJ99 A0A3B3CVH6 A0A3Q3GWC8 A0A1A7X530 A0A1B6C5J4 A0A1A8PDA8 A0A1A8JPM4 A0A1A8E856 A0A1A8HAX4 A0A1A8VH75 A0A1W4W2Y2 B4QYQ2 B4PSL3
A0A212ESR9 A0A067RKX8 D6WZI7 A0A154PG00 A0A3S2NJN3 A0A2P2I0D2 V5H2S4 A0A0P4WH28 N6TVS1 A0A1Y1KEG3 A0A195DDT3 E9IXZ0 E2B6I2 A0A158P3L8 E1ZUV5 A0A151IGW4 A0A195FUB0 A0A026WY30 A0A232F8S4 K7J7Y2 A0A2A3EKQ5 V9IID2 A0A088AIA6 A0A151WFF0 A0A1W4XHD2 A0A0C9PZW3 A0A310SHX4 A0A0M9A9G4 A0A2M4ALX5 A0A0L7RDL5 A0A1I8NNG4 A0A0K8TNG8 A0A0L0CJD8 T1E326 A0A2M3ZIS5 A0A2M4C110 A0A182FPD4 A0A1Q3EU66 A0A034W4V4 A0A0C9R9J3 W5JLE3 A0A1I8N822 U5EZE1 A0A023EIL1 A0A1L8DVV3 A0A1B0CNK8 A0A0A1WNA3 W8BI58 E9GY65 A0A0P6F0K2 A0A182P004 B0X8S5 B4M158 B4K966 A0A0M5J1E2 A0A1A9X5Y4 A0A1A9V5I4 A0A1B0ASN6 A0A1A9Z747 D3TQV5 A0A3B0KQN3 Q299A4 B4G4Y2 B3LZH3 W5USM2 A0A0K8WFW5 Q172Z9 A0A182Y5F1 A0A182M7N7 A0A182Q111 A0A182JXU4 A0A182U5U0 A0A182W1B8 A0A182XG61 Q7PQM1 A0A182RV76 A0A182IDA6 A0A084VLD7 A0A182J175 A0A3Q2PX03 B4NJ88 A0A1J1HJ99 A0A3B3CVH6 A0A3Q3GWC8 A0A1A7X530 A0A1B6C5J4 A0A1A8PDA8 A0A1A8JPM4 A0A1A8E856 A0A1A8HAX4 A0A1A8VH75 A0A1W4W2Y2 B4QYQ2 B4PSL3
PDB
4ILE
E-value=6.14413e-93,
Score=865
Ontologies
KEGG
GO
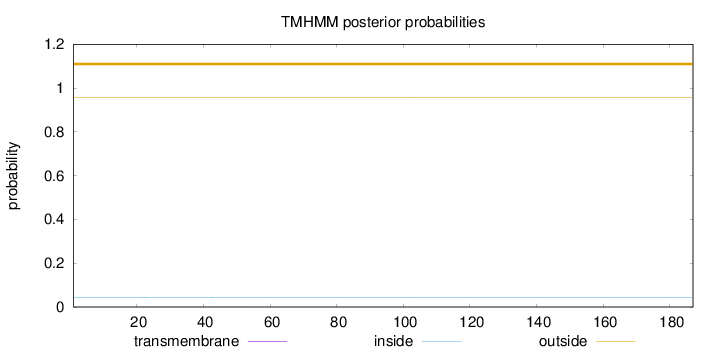
Topology
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01233
Exp number, first 60 AAs:
0.01233
Total prob of N-in:
0.04331
outside
1 - 187
Population Genetic Test Statistics
Pi
23.806494
Theta
20.517154
Tajima's D
0.463506
CLR
0.544645
CSRT
0.512774361281936
Interpretation
Uncertain
