Gene
KWMTBOMO13361
Pre Gene Modal
BGIBMGA010926
Annotation
PREDICTED:_dual_specificity_testis-specific_protein_kinase_1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.354 Nuclear Reliability : 1.663
Sequence
CDS
ATGCAACGCGAGTGTACAGAAGGATTTAGGAGTAAGTTCGAGAGGCGGAAGCCGACCAAAGTGCTAAAGATAGCAGAGAGCTTCACGAACTTGGATGAGACTGCGAAGGCTGGTCGATTTGTAAGAGGTCCGCACTCGAAGATAGGGCGTAGGTATCGCGACGGACAGGAGAGGACTAAACGCGGTGTAGTGTGCTGTGATGATGAGTGTGATAAGGAGTCAGGTGTGAGTGAAGATGGTGAAAGACTCGGTGACCGCAGAGTGACCGGCTCCTCTTGCACAGCTCTGAGAACGGCTGTGGCAGCTCTGTACCCGTTTGATGACTTCGTGCTGGAGAAAATCGGCACTGGATTCTTCTCTGAAGTTTTCAAAGTGAGTATCGCTTTGTACTAA
Protein
MQRECTEGFRSKFERRKPTKVLKIAESFTNLDETAKAGRFVRGPHSKIGRRYRDGQERTKRGVVCCDDECDKESGVSEDGERLGDRRVTGSSCTALRTAVAALYPFDDFVLEKIGTGFFSEVFKVSIALY
Summary
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF07714 Pkinase_Tyr
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
Ontologies
GO
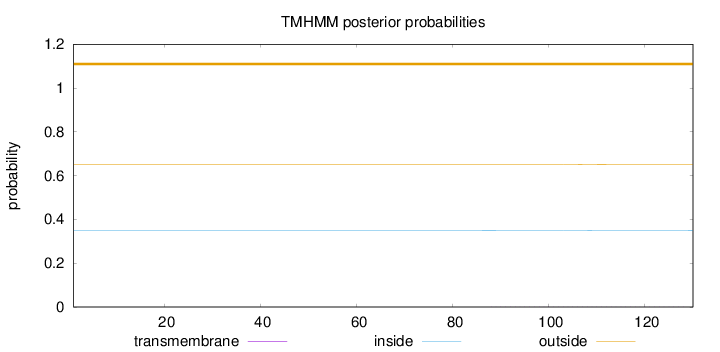
Topology
Length:
130
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02629
Exp number, first 60 AAs:
0
Total prob of N-in:
0.35022
outside
1 - 130
Population Genetic Test Statistics
Pi
199.738144
Theta
191.225932
Tajima's D
0.332222
CLR
0.182213
CSRT
0.459677016149193
Interpretation
Uncertain
