Gene
KWMTBOMO13360
Pre Gene Modal
BGIBMGA010925
Annotation
PREDICTED:_uncharacterized_protein_LOC106130174_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 2.696
Sequence
CDS
ATGCACGAAAAACACAAAAACGTGCTACAATATAAAGAGACGTTCTGCTATAGAAGAGATGCATTACCACACCTTTGCTCGCTGATCACCGGCGACGCTCTACTTTATTCCATGTTCAGAGAAAACGCTGATCCAGTCGACTGTCCTCTGAAGGCGCCTTTCTCGTTTAGCTATAATCGGGGACACGGCGATTGCAAATATCCCGTGTCGACAATAGAAAGCTGCACAGAAGACTCGAGGCTGCTCCTCAATTACCAGGCCTGTCCTGACATACACGCTTCCGAAAGTGCTGTTGAAGAACTCGAATGCTTAGCGACTTGGAAGGAGGGTAGTTTGCGGTACCTCGTAGGGAAGCTACACCACAACCACGCGACCAGCAACGAAGATAGATATAGATGCTTCGTCTATGAGAAAACAAACGGTATTGCTTCAGGTGGAAATTTGAAAGAAGATCCAATTTCGGATACTGACGTCGAGTACAGGGTTGCACAATCGGGCGATGCGACGTGCAATGGCCTGTCAAGCGCTACCGAAGGCTCCAGAACAATGGCACTAAAACGAGTGTCAGTTCGCTTCAACTGTCAATTTCCGTCGTGGATGACGTTCTCGCACACGTGGCACACACTAGACTTCACCAGCAACTACACGTTCTATCAACGCAACGCCACACTGCGGATCACGAACCAAACGGGGGCCGATATCAAAGTATACTGTGTTAACGTAAAGGCCAGTTCACCCAGCGGGAATTCTGTGGCGCTAGTTGCGCATTGGCAACATCATTGTGCATCTCGGTACGTTTGTGTTGTATTATACCGAAGAGACACATACATAGCTGAATTGCAACGTGGAGTTCCGACTTCCAAACCTGATGACGCGTGCTCCCCTCATCACTTCAACGCCGTCACGGCGCCTTACATAACTTTGGTTGCAAGCAATCCCGAATCAAAGGACTGTCCTTACCCAGGAAAATATAATATACAAAACAACAGACATCGAAGAAACGTAAGGTCTTTAGAAAGAAAAGAATATTCACATCGTAGCAGGAGCAAACGAGACGAAACTATTTTCGAACATAATCGTAGAATTAATGATAGTAGACTGTTCAATCTTAGCGTAAGAAATGTATCGAATGCTCGTATAATGAGAAGCAGGAGGCAAGCGGAGAACGCAGAACATAGTTGCGTTTTTAATAATTACAACAAACTTGAAGTTGGCTGTAATTCGGTGAAGAATATGGAGTTTTATTCGAATTGCGGGAACAAAGAAATCGTTACAGCCTACACGTGCCACGGAGGCTGGTACGAAGATGGTGCTTCATTTGTTGTGACCACCCCGGTGACCCGGGACAGCACGGCGGCCAGACGCTACTGCTTCATGTCCCGAGAGACTCGGGGCGGGACCCTGATCCTCACCCGCTCGGCAAGCAACTGCGAACGAGCTAACGTTGAGCCTGAACTCGTCTTTGATGCAACAGCTATCGGAGAGTGTCAAGATCAGACAAACGGTCAGGAAGCAGCAAGGACGTCAGGGATCCTGCTAATATGCCTGCCACTCATCGTGCTTTACATCATACCAAGATGA
Protein
MHEKHKNVLQYKETFCYRRDALPHLCSLITGDALLYSMFRENADPVDCPLKAPFSFSYNRGHGDCKYPVSTIESCTEDSRLLLNYQACPDIHASESAVEELECLATWKEGSLRYLVGKLHHNHATSNEDRYRCFVYEKTNGIASGGNLKEDPISDTDVEYRVAQSGDATCNGLSSATEGSRTMALKRVSVRFNCQFPSWMTFSHTWHTLDFTSNYTFYQRNATLRITNQTGADIKVYCVNVKASSPSGNSVALVAHWQHHCASRYVCVVLYRRDTYIAELQRGVPTSKPDDACSPHHFNAVTAPYITLVASNPESKDCPYPGKYNIQNNRHRRNVRSLERKEYSHRSRSKRDETIFEHNRRINDSRLFNLSVRNVSNARIMRSRRQAENAEHSCVFNNYNKLEVGCNSVKNMEFYSNCGNKEIVTAYTCHGGWYEDGASFVVTTPVTRDSTAARRYCFMSRETRGGTLILTRSASNCERANVEPELVFDATAIGECQDQTNGQEAARTSGILLICLPLIVLYIIPR
Summary
Uniprot
H9JN22
A0A2H1X2M0
A0A212EWH7
A0A194REJ8
A0A194Q3X9
A0A139WBC4
+ More
A0A1Y1M8S9 A0A1Y1M9F2 A0A1B6D8I4 A0A2K8JX84 A0A2J7QZJ8 A0A224XGN8 A0A1B6ML19 A0A1B6BXW0 A0A0A9XH85 A0A1B6EC96 A0A154PA97 A0A1J1HPH6 A0A0R3NLT4 A0A226D6W0 A0A1Q3FZ85 A0A3B0KI93 A0A1B6MU61 A0A2J7PZU0 T1IAU8 A0A1B6INP0 A0A1B6G0D0 A0A0B4LIF6 A0A1S4F452 A0A0P8YD76 A0A067R8C5 A0A195FWU2 A0A1I8PLI5 A0A0B4LHQ0 A0A1J1HTB4 A0A0L7QVX6 A0A1Q3FZ42 A0A1Q3FZ08 A0A1S4J286 A0A1Q3FZ90 A0A1A9WMM8 A0A1Q3FZ61 A0A1Q3FZ68 B0W206 B4R116 A0A1A9VHY2 A0A336M9N1 K7J8V1 A0A1B6D3A2 Q17FX3 E0VAJ9 A0A0M4EGN6 Q29B00 A0A2P8Y522 A0A3B0KSN4 A0A1I8MGP5 A0A1J1HNA7 B4KCU3 A0A182G485 A0A182GHD2 A0A0L7KNK1 B4PQE1 B4NKH3 A0A1B0C5Y9 A0A0T6B809 Q9VDB7 A0A0B4LHH8 B4MBG3 J9K6E3 B3LZ37 A0A2S2PLC0 B3P028 A0A1I8PLB2 T1PFE6 E0VL77 A0A2S2QJW7 A0A2K8JMZ5 B4JS46 A0A3B0K6B9 A0A336M9X3
A0A1Y1M8S9 A0A1Y1M9F2 A0A1B6D8I4 A0A2K8JX84 A0A2J7QZJ8 A0A224XGN8 A0A1B6ML19 A0A1B6BXW0 A0A0A9XH85 A0A1B6EC96 A0A154PA97 A0A1J1HPH6 A0A0R3NLT4 A0A226D6W0 A0A1Q3FZ85 A0A3B0KI93 A0A1B6MU61 A0A2J7PZU0 T1IAU8 A0A1B6INP0 A0A1B6G0D0 A0A0B4LIF6 A0A1S4F452 A0A0P8YD76 A0A067R8C5 A0A195FWU2 A0A1I8PLI5 A0A0B4LHQ0 A0A1J1HTB4 A0A0L7QVX6 A0A1Q3FZ42 A0A1Q3FZ08 A0A1S4J286 A0A1Q3FZ90 A0A1A9WMM8 A0A1Q3FZ61 A0A1Q3FZ68 B0W206 B4R116 A0A1A9VHY2 A0A336M9N1 K7J8V1 A0A1B6D3A2 Q17FX3 E0VAJ9 A0A0M4EGN6 Q29B00 A0A2P8Y522 A0A3B0KSN4 A0A1I8MGP5 A0A1J1HNA7 B4KCU3 A0A182G485 A0A182GHD2 A0A0L7KNK1 B4PQE1 B4NKH3 A0A1B0C5Y9 A0A0T6B809 Q9VDB7 A0A0B4LHH8 B4MBG3 J9K6E3 B3LZ37 A0A2S2PLC0 B3P028 A0A1I8PLB2 T1PFE6 E0VL77 A0A2S2QJW7 A0A2K8JMZ5 B4JS46 A0A3B0K6B9 A0A336M9X3
Pubmed
EMBL
BABH01030541
BABH01030542
ODYU01012522
SOQ58884.1
AGBW02011989
OWR45836.1
+ More
KQ460323 KPJ15889.1 KQ459586 KPI98115.1 KQ971372 KYB25227.1 GEZM01037084 JAV82202.1 GEZM01037085 JAV82201.1 GEDC01015326 JAS21972.1 KY031276 ATU83027.1 NEVH01009068 PNF34010.1 GFTR01007448 JAW08978.1 GEBQ01003327 JAT36650.1 GEDC01031171 JAS06127.1 GBHO01025466 GBRD01006672 GDHC01016466 JAG18138.1 JAG59149.1 JAQ02163.1 GEDC01001732 JAS35566.1 KQ434856 KZC08763.1 CVRI01000012 CRK89434.1 CM000070 KRS99829.1 LNIX01000033 OXA40598.1 GFDL01002195 JAV32850.1 OUUW01000013 SPP88220.1 GEBQ01000532 JAT39445.1 NEVH01020332 PNF21842.1 ACPB03000859 GECU01019171 JAS88535.1 GECZ01013871 JAS55898.1 AE014297 AHN57442.1 CH902617 KPU79424.1 KK852869 KDR14671.1 KQ981208 KYN44787.1 AHN57440.1 CRK89433.1 KQ414721 KOC62783.1 GFDL01002242 JAV32803.1 GFDL01002240 JAV32805.1 GFDL01002188 JAV32857.1 GFDL01002194 JAV32851.1 GFDL01002197 JAV32848.1 DS231824 EDS27442.1 CM000364 EDX12103.1 UFQT01000765 SSX27054.1 AAZX01000810 AAZX01004306 AAZX01004505 AAZX01005770 AAZX01011008 GEDC01017109 JAS20189.1 CH477267 EAT45501.2 EJY57464.1 DS235008 EEB10405.1 CP012526 ALC45334.1 EAL27199.4 PYGN01000918 PSN39357.1 SPP88221.1 CRK89435.1 CH933806 EDW16965.2 JXUM01041877 JXUM01041878 JXUM01041879 JXUM01041880 JXUM01041881 JXUM01041882 KQ561302 KXJ79063.1 JXUM01063406 JXUM01063407 KQ562247 KXJ76313.1 JTDY01007900 KOB64872.1 CM000160 EDW96250.1 CH964272 EDW85145.2 JXJN01026351 LJIG01009230 KRT83496.1 BT003233 BT004477 AAF55880.4 AAO24988.1 AAO42641.1 AHN57441.1 CH940656 EDW58434.2 ABLF02026618 ABLF02026629 EDV41911.2 GGMR01017586 MBY30205.1 CH954181 EDV48264.1 KA646633 AFP61262.1 DS235271 EEB14133.1 GGMS01008269 MBY77472.1 KY031290 ATU83041.1 CH916373 EDV94586.1 SPP88222.1 SSX27055.1
KQ460323 KPJ15889.1 KQ459586 KPI98115.1 KQ971372 KYB25227.1 GEZM01037084 JAV82202.1 GEZM01037085 JAV82201.1 GEDC01015326 JAS21972.1 KY031276 ATU83027.1 NEVH01009068 PNF34010.1 GFTR01007448 JAW08978.1 GEBQ01003327 JAT36650.1 GEDC01031171 JAS06127.1 GBHO01025466 GBRD01006672 GDHC01016466 JAG18138.1 JAG59149.1 JAQ02163.1 GEDC01001732 JAS35566.1 KQ434856 KZC08763.1 CVRI01000012 CRK89434.1 CM000070 KRS99829.1 LNIX01000033 OXA40598.1 GFDL01002195 JAV32850.1 OUUW01000013 SPP88220.1 GEBQ01000532 JAT39445.1 NEVH01020332 PNF21842.1 ACPB03000859 GECU01019171 JAS88535.1 GECZ01013871 JAS55898.1 AE014297 AHN57442.1 CH902617 KPU79424.1 KK852869 KDR14671.1 KQ981208 KYN44787.1 AHN57440.1 CRK89433.1 KQ414721 KOC62783.1 GFDL01002242 JAV32803.1 GFDL01002240 JAV32805.1 GFDL01002188 JAV32857.1 GFDL01002194 JAV32851.1 GFDL01002197 JAV32848.1 DS231824 EDS27442.1 CM000364 EDX12103.1 UFQT01000765 SSX27054.1 AAZX01000810 AAZX01004306 AAZX01004505 AAZX01005770 AAZX01011008 GEDC01017109 JAS20189.1 CH477267 EAT45501.2 EJY57464.1 DS235008 EEB10405.1 CP012526 ALC45334.1 EAL27199.4 PYGN01000918 PSN39357.1 SPP88221.1 CRK89435.1 CH933806 EDW16965.2 JXUM01041877 JXUM01041878 JXUM01041879 JXUM01041880 JXUM01041881 JXUM01041882 KQ561302 KXJ79063.1 JXUM01063406 JXUM01063407 KQ562247 KXJ76313.1 JTDY01007900 KOB64872.1 CM000160 EDW96250.1 CH964272 EDW85145.2 JXJN01026351 LJIG01009230 KRT83496.1 BT003233 BT004477 AAF55880.4 AAO24988.1 AAO42641.1 AHN57441.1 CH940656 EDW58434.2 ABLF02026618 ABLF02026629 EDV41911.2 GGMR01017586 MBY30205.1 CH954181 EDV48264.1 KA646633 AFP61262.1 DS235271 EEB14133.1 GGMS01008269 MBY77472.1 KY031290 ATU83041.1 CH916373 EDV94586.1 SPP88222.1 SSX27055.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000007266
UP000235965
+ More
UP000076502 UP000183832 UP000001819 UP000198287 UP000268350 UP000015103 UP000000803 UP000007801 UP000027135 UP000078541 UP000095300 UP000053825 UP000091820 UP000002320 UP000000304 UP000078200 UP000002358 UP000008820 UP000009046 UP000092553 UP000245037 UP000095301 UP000009192 UP000069940 UP000249989 UP000037510 UP000002282 UP000007798 UP000092460 UP000008792 UP000007819 UP000008711 UP000001070
UP000076502 UP000183832 UP000001819 UP000198287 UP000268350 UP000015103 UP000000803 UP000007801 UP000027135 UP000078541 UP000095300 UP000053825 UP000091820 UP000002320 UP000000304 UP000078200 UP000002358 UP000008820 UP000009046 UP000092553 UP000245037 UP000095301 UP000009192 UP000069940 UP000249989 UP000037510 UP000002282 UP000007798 UP000092460 UP000008792 UP000007819 UP000008711 UP000001070
Pfam
PF04193 PQ-loop
Interpro
Gene 3D
ProteinModelPortal
H9JN22
A0A2H1X2M0
A0A212EWH7
A0A194REJ8
A0A194Q3X9
A0A139WBC4
+ More
A0A1Y1M8S9 A0A1Y1M9F2 A0A1B6D8I4 A0A2K8JX84 A0A2J7QZJ8 A0A224XGN8 A0A1B6ML19 A0A1B6BXW0 A0A0A9XH85 A0A1B6EC96 A0A154PA97 A0A1J1HPH6 A0A0R3NLT4 A0A226D6W0 A0A1Q3FZ85 A0A3B0KI93 A0A1B6MU61 A0A2J7PZU0 T1IAU8 A0A1B6INP0 A0A1B6G0D0 A0A0B4LIF6 A0A1S4F452 A0A0P8YD76 A0A067R8C5 A0A195FWU2 A0A1I8PLI5 A0A0B4LHQ0 A0A1J1HTB4 A0A0L7QVX6 A0A1Q3FZ42 A0A1Q3FZ08 A0A1S4J286 A0A1Q3FZ90 A0A1A9WMM8 A0A1Q3FZ61 A0A1Q3FZ68 B0W206 B4R116 A0A1A9VHY2 A0A336M9N1 K7J8V1 A0A1B6D3A2 Q17FX3 E0VAJ9 A0A0M4EGN6 Q29B00 A0A2P8Y522 A0A3B0KSN4 A0A1I8MGP5 A0A1J1HNA7 B4KCU3 A0A182G485 A0A182GHD2 A0A0L7KNK1 B4PQE1 B4NKH3 A0A1B0C5Y9 A0A0T6B809 Q9VDB7 A0A0B4LHH8 B4MBG3 J9K6E3 B3LZ37 A0A2S2PLC0 B3P028 A0A1I8PLB2 T1PFE6 E0VL77 A0A2S2QJW7 A0A2K8JMZ5 B4JS46 A0A3B0K6B9 A0A336M9X3
A0A1Y1M8S9 A0A1Y1M9F2 A0A1B6D8I4 A0A2K8JX84 A0A2J7QZJ8 A0A224XGN8 A0A1B6ML19 A0A1B6BXW0 A0A0A9XH85 A0A1B6EC96 A0A154PA97 A0A1J1HPH6 A0A0R3NLT4 A0A226D6W0 A0A1Q3FZ85 A0A3B0KI93 A0A1B6MU61 A0A2J7PZU0 T1IAU8 A0A1B6INP0 A0A1B6G0D0 A0A0B4LIF6 A0A1S4F452 A0A0P8YD76 A0A067R8C5 A0A195FWU2 A0A1I8PLI5 A0A0B4LHQ0 A0A1J1HTB4 A0A0L7QVX6 A0A1Q3FZ42 A0A1Q3FZ08 A0A1S4J286 A0A1Q3FZ90 A0A1A9WMM8 A0A1Q3FZ61 A0A1Q3FZ68 B0W206 B4R116 A0A1A9VHY2 A0A336M9N1 K7J8V1 A0A1B6D3A2 Q17FX3 E0VAJ9 A0A0M4EGN6 Q29B00 A0A2P8Y522 A0A3B0KSN4 A0A1I8MGP5 A0A1J1HNA7 B4KCU3 A0A182G485 A0A182GHD2 A0A0L7KNK1 B4PQE1 B4NKH3 A0A1B0C5Y9 A0A0T6B809 Q9VDB7 A0A0B4LHH8 B4MBG3 J9K6E3 B3LZ37 A0A2S2PLC0 B3P028 A0A1I8PLB2 T1PFE6 E0VL77 A0A2S2QJW7 A0A2K8JMZ5 B4JS46 A0A3B0K6B9 A0A336M9X3
Ontologies
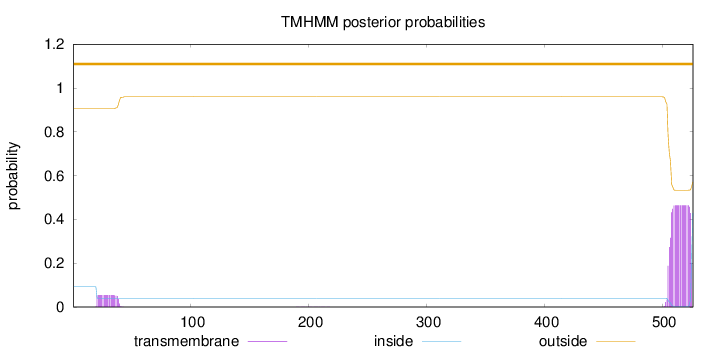
Topology
Length:
526
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.87328
Exp number, first 60 AAs:
1.02224
Total prob of N-in:
0.09362
outside
1 - 526
Population Genetic Test Statistics
Pi
24.139729
Theta
178.821058
Tajima's D
0.820931
CLR
0.380268
CSRT
0.607769611519424
Interpretation
Uncertain
