Gene
KWMTBOMO13358
Annotation
PREDICTED:_uncharacterized_protein_LOC107046166_[Diachasma_alloeum]
Location in the cell
Mitochondrial Reliability : 1.641 Nuclear Reliability : 1.716
Sequence
CDS
ATGAATGGTAGCTCTGTAGTTCTGCTGGCTACTGCTAACGTTGAAGTGCGCGACTCATCTGGTCACTTTCATACTCTTCGCGCACTCATCGACCCTGGTAGTCAATCACATTTCATTTCTAATCAAGCTATGGAACGATTGGGCTTAAAGCCTAAGCAAACTTCACGAAATGTTTGTGGCTTGGGGCAATCATCGACGTCAGTGACTGGAATGGTGGAGCTTCAACTCGGAGTTCATCAAAAGAATTTATTTAATTTAACGGCTCTCATCTTACCATCTATTTGTGCACAAATGCCCACTGCTCGTCTTGATCTTTCTTCATGCGAACACCTACGTAATTTGCGACTCGCTGATCCGAATTATAATAAGCCTGGACCAATAGATCTGCTTATCGGTGCTGAAATGTTTTCAACGCTGTTACTGCCCGGAAATATTTCAGGTAGTCCTCATGAACCCTCAGCCTTGAACACCATCTTCGGATGGATCCTGATGGGAAGTGTCAAATGCAGGGATGAGACAAAACACTCAAATTCTTTTTTTCTAAGAGAACACGATACTCCTCTCCATGAGGAATTGAGGAGACTGTGGGAGTTGGAGGTCATACCTAAATCATCAGTTCTAACGCCGGACGAGCAGCAATGTGAGAAACTTTTCACTGAAACCCACGCTCGAGGTACATCTGGAAGGTACATCGTTTCTTTGCCGTTTGGGCGGAGAGATGGGGAGTTACACTTTCCGGGTTCGCGTGAAAATACATTGCGAAGATTCCATTCATTGGAACGAAGACTTCTTTCAAGTCCAAACCTTCATCAGCAGTATTCGGACTTCATGCTGGACTACTTACATAATGGTCATAGGAGTATCCTTTTATATGCTAAGTCTCGCGTGGCCCCAATTAAGAGAGTTTCATTGCCGAGACTAGAGCTCTGCGCTGCGGTTCTGCTCGCTGACCTGCTCAAATTCGTAAAAGAAGTGTATGTACCGTTACTTCCGCTTTCTGATACATTTGCTTGGTCAGATTCAACGGTAACTCTGTCATGGATTCGTGGCCATTCCTTGCGTTGGAAAACATTTGTTGCTAATAGGGTTAGTCATATCCAAGAAACTGTCCCCGAAACTCACTGGAATCACGTGCGCTCTAATGAAAATCCTGCGGACATAGCTTCCCGCGGTATGTGTCCGAGCGACCTACTCCAGAGTGAGTTATGGTGGGCGGGACCGAGCTGGCTTCAGCAACCTTCATCTTCATGGCCTTCAAATTCCGGTCTCAAGGTGAACGAGACTGATATGCTTACCGAGGAACGCTGTTTTTCGTTGATCGTGAATGATGAACCTAACTTTATCGATTCTTTGCTCGAAAGGTTTTCATCAATTCGTAAGGTTCAACGTATTATTGGTTATTGTCTTCGTTTTATCAGAAATGCTAACAAAAAACAGACACGTGTTCCGGGTACCTTTTTAGGTCGTGCAGAACTTCATGGGGCTCTGATGACGATTACCAAGCGAATTCAATTTCTCAACTTTCAGGATGAAATTCGTAAGCTCGAGAATGGTATACCATTACCCAAACCGTGGAGAAAACTCAATCCATTCCTGGACGACCAAGGAGTTGTCAGGGTGGGCGGCCGTTTGTCTCGTTCTGGTCTTGAGTTTAGTCATAAGCATCCTGCTCTGTTGCCGCGTCATTCTGGGTTCACATACATGATCATCGAAGCAATTCACAAAGAGAATTGTCATCCTGGGTTGGGAACGACACAATATCTTTTGATGCAGCTGTTTTGGAGAGCAAAGTGGAACAAAAGGGGCGAAACTTTGAATGATGGTTCTGTTGTACTGATTAAGAGTGAAAGTACAATGCCGTTGCACTGGCCTTTAGCTCGTGTCTCCAAGCTTCACGCCAGCCCTGATGGCGTGGTTCGTATTGCCACTGTCCGTACAGCCAATGGCAAATTCTACACTAGGCCATTAACGAAGTTAGCTCCCCTACCTATTTATTCTGATTAA
Protein
MNGSSVVLLATANVEVRDSSGHFHTLRALIDPGSQSHFISNQAMERLGLKPKQTSRNVCGLGQSSTSVTGMVELQLGVHQKNLFNLTALILPSICAQMPTARLDLSSCEHLRNLRLADPNYNKPGPIDLLIGAEMFSTLLLPGNISGSPHEPSALNTIFGWILMGSVKCRDETKHSNSFFLREHDTPLHEELRRLWELEVIPKSSVLTPDEQQCEKLFTETHARGTSGRYIVSLPFGRRDGELHFPGSRENTLRRFHSLERRLLSSPNLHQQYSDFMLDYLHNGHRSILLYAKSRVAPIKRVSLPRLELCAAVLLADLLKFVKEVYVPLLPLSDTFAWSDSTVTLSWIRGHSLRWKTFVANRVSHIQETVPETHWNHVRSNENPADIASRGMCPSDLLQSELWWAGPSWLQQPSSSWPSNSGLKVNETDMLTEERCFSLIVNDEPNFIDSLLERFSSIRKVQRIIGYCLRFIRNANKKQTRVPGTFLGRAELHGALMTITKRIQFLNFQDEIRKLENGIPLPKPWRKLNPFLDDQGVVRVGGRLSRSGLEFSHKHPALLPRHSGFTYMIIEAIHKENCHPGLGTTQYLLMQLFWRAKWNKRGETLNDGSVVLIKSESTMPLHWPLARVSKLHASPDGVVRIATVRTANGKFYTRPLTKLAPLPIYSD
Summary
Uniprot
EMBL
Proteomes
Interpro
SUPFAM
SSF53098
SSF53098
Gene 3D
ProteinModelPortal
Ontologies
KEGG
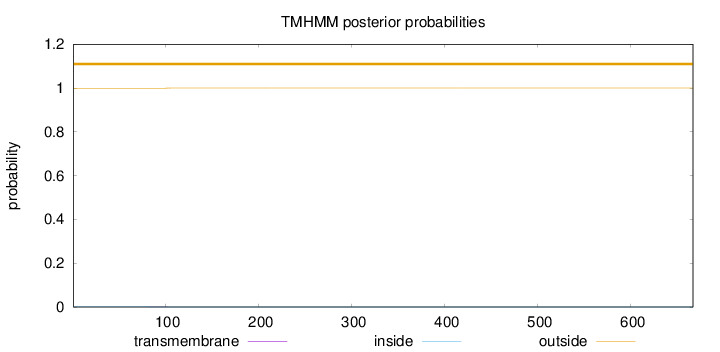
Topology
Length:
667
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0143199999999999
Exp number, first 60 AAs:
0.00013
Total prob of N-in:
0.00077
outside
1 - 667
Population Genetic Test Statistics
Pi
391.353879
Theta
175.967369
Tajima's D
3.267805
CLR
0.204242
CSRT
0.990300484975751
Interpretation
Uncertain
