Gene
KWMTBOMO13357
Annotation
PREDICTED:_elongation_of_very_long_chain_fatty_acids_protein_4-like_isoform_X2_[Bombyx_mori]
Full name
Elongation of very long chain fatty acids protein
+ More
Elongation of very long chain fatty acids protein 4
Elongation of very long chain fatty acids protein 4
Alternative Name
Very-long-chain 3-oxoacyl-CoA synthase
3-keto acyl-CoA synthase ELOVL4
ELOVL fatty acid elongase 4
Very long chain 3-ketoacyl-CoA synthase 4
Very long chain 3-oxoacyl-CoA synthase 4
3-keto acyl-CoA synthase ELOVL4
ELOVL fatty acid elongase 4
Very long chain 3-ketoacyl-CoA synthase 4
Very long chain 3-oxoacyl-CoA synthase 4
Location in the cell
PlasmaMembrane Reliability : 3.475
Sequence
CDS
ATGAAGCAACGCGAACCTTACAAACTGAAGAGCTTGCTTTTGTTTTACAACTGTTTTCAAGTCGCTTTCTCTGCTTATTTGGTTTTTATTTATTCCAGGTATATTCTAAGTTACGGTGTGATCACAAAAAGATGTCCCAAAGGTGAAGATTTACAAGCGGTTATCAAAGAAATCTATCCATATTTCATAGCAAAGCACCTTGATCTACTAGATACAGTGTTCTTCGTTCTGAGAAAGAAAGACAACCAAGTGACTTTCCTTCATCTTTACCACCACAACACCATGGTGACTTGGACCTGGCTCCATTTAATGTACCATCCCGCAGATCACTTTGTGGTGGTTGGCATGTTGAATAGCTTCGTTCATGTGCTGATGTATGCTTATTATGGGATATCGTCTCTGGGCCCGAAGTATGCTAAGTTTGTTTGGTGGAAGAAACATTTGACTAAGGTGCAACTGGTGCAGTTCGTCTTAGTTACTTCCCACCTGCACTACCAGCAGAAGTTGACGCCTTGTCCTCTACCAACGTTTTTCCATTACTTCTGCGTGTCACTTATTTGCTCCTTTTTCTTATTGTTCATGAAATTCTACGTTCAGAGCTACAGAGTTAGAAAGGAGAGGAAAGCGAACGATTTAAATGTGGGAAATAAGTTAGGCGGGGAGCGATTGTTAAAGGGACTTTGA
Protein
MKQREPYKLKSLLLFYNCFQVAFSAYLVFIYSRYILSYGVITKRCPKGEDLQAVIKEIYPYFIAKHLDLLDTVFFVLRKKDNQVTFLHLYHHNTMVTWTWLHLMYHPADHFVVVGMLNSFVHVLMYAYYGISSLGPKYAKFVWWKKHLTKVQLVQFVLVTSHLHYQQKLTPCPLPTFFHYFCVSLICSFFLLFMKFYVQSYRVRKERKANDLNVGNKLGGERLLKGL
Summary
Description
Catalyzes the first and rate-limiting reaction of the four reactions that constitute the long-chain fatty acids elongation cycle. This endoplasmic reticulum-bound enzymatic process allows the addition of 2 carbons to the chain of long- and very long-chain fatty acids (VLCFAs) per cycle. Condensing enzyme that specifically elongates C24:0 and C26:0 acyl-CoAs. May participate to the production of saturated and monounsaturated VLCFAs of different chain lengths that are involved in multiple biological processes as precursors of membrane lipids and lipid mediators. May play a critical role in early brain and skin development.
Catalytic Activity
a very-long-chain acyl-CoA + H(+) + malonyl-CoA = a very-long-chain 3-oxoacyl-CoA + CO2 + CoA
Subunit
Oligomer.
Similarity
Belongs to the ELO family.
Belongs to the ELO family. ELOVL4 subfamily.
Belongs to the ELO family. ELOVL4 subfamily.
Uniprot
A0A2H1VZH1
A0A2A4JNZ7
A0A2A4IT40
A0A212EWG2
A0A194Q3Y4
A0A194RIW9
+ More
A0A194RE66 A0A194PZF1 A0A212EWL1 A0A194PXS2 A0A2H1X1Z6 A0A194REJ3 T1PEI5 A0A1E1WEQ6 B4JF47 A0A1I8NVN9 A0A1W4X4I5 B3P8N4 A0A1E1WBM7 A0A1W4WV07 B4R0M2 B4HEP6 A0A2A4IYB4 A0A194PYL9 B4K959 B4PMB5 Q9VCY7 B3M000 C6SV14 A0A336MEX6 A0A0A1XAS6 A0A0M4F374 A0A2H1WW52 B4M164 A0A1A9V750 A0A1A9Z486 W8AWR6 A0A1A9XJ22 A0A1B0BXZ2 A0A1Y1LS48 A0A2A4IXM4 K1R6N6 B4NJS5 A0A336M748 Q29BK4 A0A1W4VR29 B4GP37 A0A1A9W5Z6 A0A3B0JF55 A0A194RDG5 A0A336KLJ3 A0A3B0JEF4 A0A2H1VQJ9 A0A2A4J2V1 A0A1W4WV09 E0VAZ6 F1RQL1 A0A069DYC2 T1HHY5 A0A336L063 A0A084VMT0 A0A034WG82 A0A287A8V2 A0A034WKC9 A0A182Q5E2 A0A075QRX8 A0A0K8VXE8 A0A0L0CGV6 I3LVJ0 A0A336K2J4 A0A1B0FQK1 A0A023F182 A0A182F0V8 A0A2H1X1Z5 A0A194PYM4 H9JN16 A0A158NN74 H3A2B9 E0VQR5 A0A154PBW3 A0A1I8NHJ7 H9JN18 W5J2I2 A0A1W2W9A1 H9JN19 A0A151I3K9 H0X3V6 A0A1B0D768 A0A2H1WH44 A0A2R7WMF5 A0A182IU15 G3WRL1 A0A336M016 A0A1S3MP43 A0A2A4IWR2 S4PNY7 A0A060WDY9 H9JN15 A0A158NS34 A0A091MTW9
A0A194RE66 A0A194PZF1 A0A212EWL1 A0A194PXS2 A0A2H1X1Z6 A0A194REJ3 T1PEI5 A0A1E1WEQ6 B4JF47 A0A1I8NVN9 A0A1W4X4I5 B3P8N4 A0A1E1WBM7 A0A1W4WV07 B4R0M2 B4HEP6 A0A2A4IYB4 A0A194PYL9 B4K959 B4PMB5 Q9VCY7 B3M000 C6SV14 A0A336MEX6 A0A0A1XAS6 A0A0M4F374 A0A2H1WW52 B4M164 A0A1A9V750 A0A1A9Z486 W8AWR6 A0A1A9XJ22 A0A1B0BXZ2 A0A1Y1LS48 A0A2A4IXM4 K1R6N6 B4NJS5 A0A336M748 Q29BK4 A0A1W4VR29 B4GP37 A0A1A9W5Z6 A0A3B0JF55 A0A194RDG5 A0A336KLJ3 A0A3B0JEF4 A0A2H1VQJ9 A0A2A4J2V1 A0A1W4WV09 E0VAZ6 F1RQL1 A0A069DYC2 T1HHY5 A0A336L063 A0A084VMT0 A0A034WG82 A0A287A8V2 A0A034WKC9 A0A182Q5E2 A0A075QRX8 A0A0K8VXE8 A0A0L0CGV6 I3LVJ0 A0A336K2J4 A0A1B0FQK1 A0A023F182 A0A182F0V8 A0A2H1X1Z5 A0A194PYM4 H9JN16 A0A158NN74 H3A2B9 E0VQR5 A0A154PBW3 A0A1I8NHJ7 H9JN18 W5J2I2 A0A1W2W9A1 H9JN19 A0A151I3K9 H0X3V6 A0A1B0D768 A0A2H1WH44 A0A2R7WMF5 A0A182IU15 G3WRL1 A0A336M016 A0A1S3MP43 A0A2A4IWR2 S4PNY7 A0A060WDY9 H9JN15 A0A158NS34 A0A091MTW9
EC Number
2.3.1.199
Pubmed
22118469
26354079
25315136
17994087
18057021
17550304
+ More
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 24495485 28004739 22992520 15632085 23185243 20566863 26334808 24438588 25348373 24970595 26108605 25474469 19121390 21347285 9215903 20920257 23761445 21709235 23622113 24755649
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 24495485 28004739 22992520 15632085 23185243 20566863 26334808 24438588 25348373 24970595 26108605 25474469 19121390 21347285 9215903 20920257 23761445 21709235 23622113 24755649
EMBL
ODYU01005098
SOQ45634.1
NWSH01000869
PCG73785.1
NWSH01008171
PCG62666.1
+ More
AGBW02011990 OWR45832.1 KQ459586 KPI98120.1 KQ460323 KPJ15886.1 KPJ15887.1 KPI98119.1 OWR45834.1 KPI98122.1 ODYU01012860 SOQ59370.1 KPJ15884.1 KA647151 AFP61780.1 GDQN01005618 JAT85436.1 CH916369 EDV93328.1 CH954182 EDV54129.1 GDQN01006662 JAT84392.1 CM000364 EDX13945.1 CH480815 EDW43207.1 NWSH01005494 PCG64160.1 KPI98123.1 CH933806 EDW14472.1 KRG01026.1 CM000160 EDW98020.1 AE014297 AY061182 AAF56018.1 AAL28730.1 CH902617 EDV44190.1 KPU80676.1 BT089006 ACT88147.1 UFQT01000742 SSX26877.1 GBXI01008419 GBXI01006261 JAD05873.1 JAD08031.1 CP012526 ALC45632.1 ODYU01011454 SOQ57196.1 CH940650 EDW67475.1 KRF83305.1 GAMC01021294 GAMC01021292 JAB85261.1 JXJN01022417 GEZM01052596 JAV74396.1 NWSH01004932 PCG64575.1 JH818916 EKC39214.1 CH964272 EDW85037.1 UFQT01000651 SSX26132.1 CM000070 EAL26992.1 CH479186 EDW38920.1 OUUW01000005 SPP80765.1 KPJ15883.1 UFQS01000651 SSX05774.1 SSX26133.1 SPP80764.1 ODYU01003837 SOQ43090.1 NWSH01003488 PCG66199.1 DS235019 EEB10552.1 AEMK02000001 GBGD01002495 JAC86394.1 ACPB03001018 UFQS01000742 SSX06530.1 SSX26879.1 ATLV01014624 KE524975 KFB39274.1 GAKP01004356 JAC54596.1 GAKP01004357 JAC54595.1 AXCN02000640 KF964010 AIG21331.1 GDHF01008780 JAI43534.1 JRES01000413 KNC31471.1 UFQS01000065 UFQT01000065 SSW98909.1 SSX19295.1 CCAG010007298 CCAG010007299 GBBI01003435 JAC15277.1 ODYU01012850 SOQ59355.1 KPI98128.1 BABH01030565 BABH01030566 ADTU01021114 ADTU01021115 AFYH01242903 AFYH01242904 AFYH01242905 AFYH01242906 AFYH01242907 AFYH01242908 AFYH01242909 AFYH01242910 AFYH01242911 DS235435 EEB15720.1 KQ434856 KZC08758.1 BABH01030564 ADMH02002132 ETN58472.1 BABH01030562 KQ976475 KYM83922.1 AAQR03130013 AAQR03130014 AJVK01012348 AJVK01012349 ODYU01008314 SOQ51794.1 KK854897 PTY19585.1 AEFK01168560 AEFK01168561 AEFK01168562 SSX19298.1 PCG64161.1 GAIX01000552 JAA92008.1 FR904503 CDQ65367.1 BABH01030569 ADTU01024445 ADTU01024446 ADTU01024447 ADTU01024448 ADTU01024449 ADTU01024450 ADTU01024451 ADTU01024452 KK516525 KFP64935.1
AGBW02011990 OWR45832.1 KQ459586 KPI98120.1 KQ460323 KPJ15886.1 KPJ15887.1 KPI98119.1 OWR45834.1 KPI98122.1 ODYU01012860 SOQ59370.1 KPJ15884.1 KA647151 AFP61780.1 GDQN01005618 JAT85436.1 CH916369 EDV93328.1 CH954182 EDV54129.1 GDQN01006662 JAT84392.1 CM000364 EDX13945.1 CH480815 EDW43207.1 NWSH01005494 PCG64160.1 KPI98123.1 CH933806 EDW14472.1 KRG01026.1 CM000160 EDW98020.1 AE014297 AY061182 AAF56018.1 AAL28730.1 CH902617 EDV44190.1 KPU80676.1 BT089006 ACT88147.1 UFQT01000742 SSX26877.1 GBXI01008419 GBXI01006261 JAD05873.1 JAD08031.1 CP012526 ALC45632.1 ODYU01011454 SOQ57196.1 CH940650 EDW67475.1 KRF83305.1 GAMC01021294 GAMC01021292 JAB85261.1 JXJN01022417 GEZM01052596 JAV74396.1 NWSH01004932 PCG64575.1 JH818916 EKC39214.1 CH964272 EDW85037.1 UFQT01000651 SSX26132.1 CM000070 EAL26992.1 CH479186 EDW38920.1 OUUW01000005 SPP80765.1 KPJ15883.1 UFQS01000651 SSX05774.1 SSX26133.1 SPP80764.1 ODYU01003837 SOQ43090.1 NWSH01003488 PCG66199.1 DS235019 EEB10552.1 AEMK02000001 GBGD01002495 JAC86394.1 ACPB03001018 UFQS01000742 SSX06530.1 SSX26879.1 ATLV01014624 KE524975 KFB39274.1 GAKP01004356 JAC54596.1 GAKP01004357 JAC54595.1 AXCN02000640 KF964010 AIG21331.1 GDHF01008780 JAI43534.1 JRES01000413 KNC31471.1 UFQS01000065 UFQT01000065 SSW98909.1 SSX19295.1 CCAG010007298 CCAG010007299 GBBI01003435 JAC15277.1 ODYU01012850 SOQ59355.1 KPI98128.1 BABH01030565 BABH01030566 ADTU01021114 ADTU01021115 AFYH01242903 AFYH01242904 AFYH01242905 AFYH01242906 AFYH01242907 AFYH01242908 AFYH01242909 AFYH01242910 AFYH01242911 DS235435 EEB15720.1 KQ434856 KZC08758.1 BABH01030564 ADMH02002132 ETN58472.1 BABH01030562 KQ976475 KYM83922.1 AAQR03130013 AAQR03130014 AJVK01012348 AJVK01012349 ODYU01008314 SOQ51794.1 KK854897 PTY19585.1 AEFK01168560 AEFK01168561 AEFK01168562 SSX19298.1 PCG64161.1 GAIX01000552 JAA92008.1 FR904503 CDQ65367.1 BABH01030569 ADTU01024445 ADTU01024446 ADTU01024447 ADTU01024448 ADTU01024449 ADTU01024450 ADTU01024451 ADTU01024452 KK516525 KFP64935.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000095301
UP000001070
+ More
UP000095300 UP000192223 UP000008711 UP000000304 UP000001292 UP000009192 UP000002282 UP000000803 UP000007801 UP000092553 UP000008792 UP000078200 UP000092445 UP000092443 UP000092460 UP000005408 UP000007798 UP000001819 UP000192221 UP000008744 UP000091820 UP000268350 UP000009046 UP000008227 UP000015103 UP000030765 UP000075886 UP000037069 UP000092444 UP000069272 UP000005204 UP000005205 UP000008672 UP000076502 UP000000673 UP000078540 UP000005225 UP000092462 UP000075880 UP000007648 UP000087266 UP000193380
UP000095300 UP000192223 UP000008711 UP000000304 UP000001292 UP000009192 UP000002282 UP000000803 UP000007801 UP000092553 UP000008792 UP000078200 UP000092445 UP000092443 UP000092460 UP000005408 UP000007798 UP000001819 UP000192221 UP000008744 UP000091820 UP000268350 UP000009046 UP000008227 UP000015103 UP000030765 UP000075886 UP000037069 UP000092444 UP000069272 UP000005204 UP000005205 UP000008672 UP000076502 UP000000673 UP000078540 UP000005225 UP000092462 UP000075880 UP000007648 UP000087266 UP000193380
Pfam
PF01151 ELO
ProteinModelPortal
A0A2H1VZH1
A0A2A4JNZ7
A0A2A4IT40
A0A212EWG2
A0A194Q3Y4
A0A194RIW9
+ More
A0A194RE66 A0A194PZF1 A0A212EWL1 A0A194PXS2 A0A2H1X1Z6 A0A194REJ3 T1PEI5 A0A1E1WEQ6 B4JF47 A0A1I8NVN9 A0A1W4X4I5 B3P8N4 A0A1E1WBM7 A0A1W4WV07 B4R0M2 B4HEP6 A0A2A4IYB4 A0A194PYL9 B4K959 B4PMB5 Q9VCY7 B3M000 C6SV14 A0A336MEX6 A0A0A1XAS6 A0A0M4F374 A0A2H1WW52 B4M164 A0A1A9V750 A0A1A9Z486 W8AWR6 A0A1A9XJ22 A0A1B0BXZ2 A0A1Y1LS48 A0A2A4IXM4 K1R6N6 B4NJS5 A0A336M748 Q29BK4 A0A1W4VR29 B4GP37 A0A1A9W5Z6 A0A3B0JF55 A0A194RDG5 A0A336KLJ3 A0A3B0JEF4 A0A2H1VQJ9 A0A2A4J2V1 A0A1W4WV09 E0VAZ6 F1RQL1 A0A069DYC2 T1HHY5 A0A336L063 A0A084VMT0 A0A034WG82 A0A287A8V2 A0A034WKC9 A0A182Q5E2 A0A075QRX8 A0A0K8VXE8 A0A0L0CGV6 I3LVJ0 A0A336K2J4 A0A1B0FQK1 A0A023F182 A0A182F0V8 A0A2H1X1Z5 A0A194PYM4 H9JN16 A0A158NN74 H3A2B9 E0VQR5 A0A154PBW3 A0A1I8NHJ7 H9JN18 W5J2I2 A0A1W2W9A1 H9JN19 A0A151I3K9 H0X3V6 A0A1B0D768 A0A2H1WH44 A0A2R7WMF5 A0A182IU15 G3WRL1 A0A336M016 A0A1S3MP43 A0A2A4IWR2 S4PNY7 A0A060WDY9 H9JN15 A0A158NS34 A0A091MTW9
A0A194RE66 A0A194PZF1 A0A212EWL1 A0A194PXS2 A0A2H1X1Z6 A0A194REJ3 T1PEI5 A0A1E1WEQ6 B4JF47 A0A1I8NVN9 A0A1W4X4I5 B3P8N4 A0A1E1WBM7 A0A1W4WV07 B4R0M2 B4HEP6 A0A2A4IYB4 A0A194PYL9 B4K959 B4PMB5 Q9VCY7 B3M000 C6SV14 A0A336MEX6 A0A0A1XAS6 A0A0M4F374 A0A2H1WW52 B4M164 A0A1A9V750 A0A1A9Z486 W8AWR6 A0A1A9XJ22 A0A1B0BXZ2 A0A1Y1LS48 A0A2A4IXM4 K1R6N6 B4NJS5 A0A336M748 Q29BK4 A0A1W4VR29 B4GP37 A0A1A9W5Z6 A0A3B0JF55 A0A194RDG5 A0A336KLJ3 A0A3B0JEF4 A0A2H1VQJ9 A0A2A4J2V1 A0A1W4WV09 E0VAZ6 F1RQL1 A0A069DYC2 T1HHY5 A0A336L063 A0A084VMT0 A0A034WG82 A0A287A8V2 A0A034WKC9 A0A182Q5E2 A0A075QRX8 A0A0K8VXE8 A0A0L0CGV6 I3LVJ0 A0A336K2J4 A0A1B0FQK1 A0A023F182 A0A182F0V8 A0A2H1X1Z5 A0A194PYM4 H9JN16 A0A158NN74 H3A2B9 E0VQR5 A0A154PBW3 A0A1I8NHJ7 H9JN18 W5J2I2 A0A1W2W9A1 H9JN19 A0A151I3K9 H0X3V6 A0A1B0D768 A0A2H1WH44 A0A2R7WMF5 A0A182IU15 G3WRL1 A0A336M016 A0A1S3MP43 A0A2A4IWR2 S4PNY7 A0A060WDY9 H9JN15 A0A158NS34 A0A091MTW9
Ontologies
PATHWAY
GO
GO:0102338
GO:0016021
GO:0102756
GO:0102337
GO:0006633
GO:0102336
GO:0007110
GO:0007112
GO:0042761
GO:0051225
GO:0009922
GO:0000212
GO:0030497
GO:0042811
GO:0000915
GO:0007111
GO:0034626
GO:0030148
GO:0019367
GO:0030176
GO:0034625
GO:0006636
GO:0035338
GO:0006508
GO:0007017
GO:0005200
GO:0005525
GO:0031386
GO:0048869
GO:0003676
GO:0016491
GO:0015930
PANTHER
Topology
Subcellular location
Endoplasmic reticulum membrane
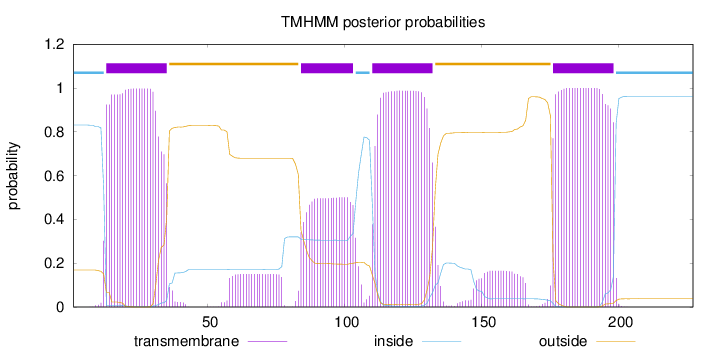
Length:
227
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
82.76128
Exp number, first 60 AAs:
22.40638
Total prob of N-in:
0.83110
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 83
TMhelix
84 - 103
inside
104 - 109
TMhelix
110 - 132
outside
133 - 175
TMhelix
176 - 198
inside
199 - 227
Population Genetic Test Statistics
Pi
182.676473
Theta
172.920545
Tajima's D
-0.545287
CLR
129.557249
CSRT
0.235438228088596
Interpretation
Uncertain
