Gene
KWMTBOMO13343
Annotation
PREDICTED:_uncharacterized_protein_LOC101737183_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.084 Mitochondrial Reliability : 1.488
Sequence
CDS
ATGCACACGAGGTTGGCTGAGGCGGGAAAATTGCTGGAACGTATTCTGGCTGCCCGCATCGTTCGGCACCTGGTCGGGGTGGGGCCTGACCTGTCGGCGAAGCAGTACGGCTTCCGGGAGGGCCGTTCGACCGTGGATGCAATTCGTCGCGTGCGGTCCCTCTCGGATGAGGCCGTTTCTCGGGGTGGGGTGGCGCTGGCGGTGTCTCTTGACATCGCCAACGCATTTAACACTCTGCCCTGGACCGTGATAGGGGGGGCACTAGAGAGGCATGGAGTGCCCCTCTACCTCCGCCGGCTGGTTGGGTCCTATCTGGGGGCCAGGTCGGTCGTATGTACCGGGTACGGTGGGACCCTTCATCGTTTTCCGGTCGTGCGTGGTGTTCCGCAGGGGTCGGTTCTCGGCCCCCTCTTGTGGAATATCGGGTACGACTGGGTGCTGAGGGGCGCCCTCCTCCCGGGCCTCCGCGTTATTTGTTACGCGGACGACACGTTGGTCGTGGCCCGGGGGGATGATTATAGGGAGTCTGCCCGTCTCGCCACGGCGGGAGTGGCCCTCGTCGTCGGAAGGATAAGGAGGCTGGGTCTCGACGTGGCGCTCAGTAAATCCGAGGCTCTGTGGTTTCACGGGCCGCGGAGGGCGCCACCCGTTGACACCCACATCGTGGTTGGAGGCGTCCGGATAGGGGCGGCGTGTGTTTTGGTGGGGACGCCGCCATGGGAGCTGGAGGCGGAGTCGCTCGCTGCCGACTATCGGTGGCGCAGCGAGCTTCGTGCTCGGGGCGTGGCGCGTCTCCTCGAAAGTGAGCTGCGGGCGCGGAAGGCCCATTCTCGGCGGTCCGTGCTCGAGTCGTGGTCGAGGCGATTGGCCAACCCCACGTGGGGGCTACGGACCGTCGAGGCGGTTTACCCGGTCTTTGATGACTGGGTGAATCGTGGCGAAGGACGTCTCACCTTTCGTCTGGTGCAGGTGCTGACCGGGCACGGATGCTTCGGGAAGTACCTGCGCCGGATAGGGGCTGAGCCGATGACGAGGTGTCACCATTGTGGACACGACCTGGAAACGGCGGAGCATACGCTCGCTGTCTGCCCTGCGTGGGAGGTGCAGCGCCGTGTCCGGGTCGCAAAGATAGGACCTGACTTGTCGCTGCCTGGCGTCGTGGCGTCGATGCTTGGCAGCGATGACTCATGGAAGGCTATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGGGCGAGTGAGGGAAAGCTCTCCTCACTACGCAGAAACCCGCCGCCGCCGAGCAGGGGGTCGGGACCGGGGTCGTATCCGTGACCTGGCCCCCTAA
Protein
MHTRLAEAGKLLERILAARIVRHLVGVGPDLSAKQYGFREGRSTVDAIRRVRSLSDEAVSRGGVALAVSLDIANAFNTLPWTVIGGALERHGVPLYLRRLVGSYLGARSVVCTGYGGTLHRFPVVRGVPQGSVLGPLLWNIGYDWVLRGALLPGLRVICYADDTLVVARGDDYRESARLATAGVALVVGRIRRLGLDVALSKSEALWFHGPRRAPPVDTHIVVGGVRIGAACVLVGTPPWELEAESLAADYRWRSELRARGVARLLESELRARKAHSRRSVLESWSRRLANPTWGLRTVEAVYPVFDDWVNRGEGRLTFRLVQVLTGHGCFGKYLRRIGAEPMTRCHHCGHDLETAEHTLAVCPAWEVQRRVRVAKIGPDLSLPGVVASMLGSDDSWKAMLDFCECTISQKEAAGRVRESSPHYAETRRRRAGGRDRGRIRDLAP
Summary
Uniprot
EMBL
RSAL01003504
RVE40273.1
LBMM01003679
KMQ93242.1
RSAL01000109
RVE47213.1
+ More
LBMM01004906 KMQ92001.1 LBMM01004603 KMQ92250.1 LBMM01004344 KMQ92531.1 LBMM01008162 KMQ89092.1 LBMM01006110 KMQ90925.1 LBMM01006662 KMQ90413.1 LBMM01006977 KMQ90149.1 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 ABLF02016340 ABLF02038790 ABLF02038792 ABLF02021645
LBMM01004906 KMQ92001.1 LBMM01004603 KMQ92250.1 LBMM01004344 KMQ92531.1 LBMM01008162 KMQ89092.1 LBMM01006110 KMQ90925.1 LBMM01006662 KMQ90413.1 LBMM01006977 KMQ90149.1 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 ABLF02016340 ABLF02038790 ABLF02038792 ABLF02021645
Proteomes
Interpro
Gene 3D
ProteinModelPortal
PDB
6AR3
E-value=3.8372e-06,
Score=121
Ontologies
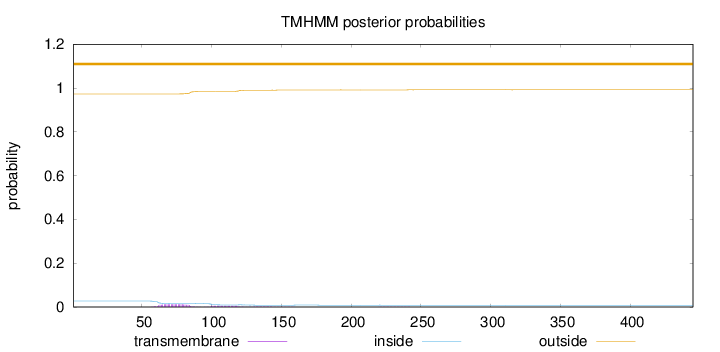
Topology
Length:
445
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.44748
Exp number, first 60 AAs:
0.0085
Total prob of N-in:
0.02715
outside
1 - 445
Population Genetic Test Statistics
Pi
30.300735
Theta
45.576865
Tajima's D
-1.137183
CLR
0.113956
CSRT
0.113944302784861
Interpretation
Uncertain
