Pre Gene Modal
BGIBMGA010951
Annotation
PREDICTED:_exocyst_complex_component_4_[Bombyx_mori]
Full name
Exocyst complex component 4
Alternative Name
Exocyst complex component Sec8
Location in the cell
Mitochondrial Reliability : 1.872
Sequence
CDS
ATGTCCTCTCCTCCACCAACGAAACCACCCCGTGGGGTTAAGCAAGGAAAAGAAACGAGTGGCCTGCTAATGTCGGTGATCAGAACACTGTCAGCGAGTGAGACCAATGAACAACGTGAAAAGGAACGCGCTAAACTTGAGAAAGAGTACAAGAAGAGCGATGCAAGGCTGGATGAGCTAGTTGCTGTTCATGCACAGGAGATTACGGATGTTATGCAAAAATTCACAGCTGTGGGTACAGCCTTATCAGTATGGGGACAGCATTGTGAGGCAGCAGTGGCTCGTCTGGGCGCATGCCGAGGTTTGTTACGATTAAGACGTGATGACCTAAGAAGATTGTGGGCGGATGCAAGAACCCATCACTATGCACTACAGATGTTAACTGATATAGAACGTGTAGTAGTTTCGGAACGTGAAACTCGCGAATCGTTATCAGCTGGTCGTGTACTAGCTGCAGCTTATACATTGAAAGCTGCCCTAGATACAGCAGACACTACACTGTCCCATGTTGATGCATTAAACACCACAAGGCAGCATTTGCAGGCAAAAAAACGTGAACTGGTAGATGTCATTGTTCGTCGAGTGAATGATGCTGTCTATCGTGGCGAAACAGCACCAATGACGAGAAGACTATCTGCTAGAAGGCGTACAGCAATCCTGTTGGCCGAACTGACAAAGAATGAAGAAGTGACTGATGCTTATCTACAAGATATCACCCCTGAGTACGAAGCGTCACTTTCTGAAGATGACAAAACATTCGAGAGTACTGTTATGATATCACTAGAAGCCTTAGGAGTATTGGAACAACTGAAGGAAGCTACTGAGAAATTCAAGGTGCAAATTCAAAATGAATTATTAGAGGTCATCGATTCGGTCTCTCGCCGAATCATCGATGAAGGCGATGTAGAAATTGACCAGGAGTGTGACGAGGATGGCGAGACACAAGAAGCCGAGGGAGTGACAGAGGCCGGGAGACCACTCGCCAGACTGGTCGCTAGTCTGGGAGAAGAGCTTAGGGCGTGCGCTGCAAGAAATAAGAGGCTTTTGCAGTTATGGCGAGGTGCCTTACAGAAACATCGACTGTCTGATGCAACTCTGCACACAGAACAACACTACTGGAGTGCCGTACAACAAGTTCTTCAACTTCTGCTGACTGAATACTTGGAGATAGAGAGCGTGAACTTAGCAGCTCAGCGCTCTGGCACTACCACAGTAGAAGCGGCCGGTTCCCAACTACGTCTGACCGACTACTTCTCCAAGAAACGACCTCAGAGGCAACACACCAAACTCTTCAGATTTACAGGCCCTACCGCAGTGACGCCCAGTCTGCAGGCTCGCCGGCACGGGTACCCGCTGGTGTGCCGCGCCGACCCGGCTCATCTGCACGCGGTGTTGCCAGCTCTGCACGCCTTGTGTGCCGACATCGAACCTCTTACAGGTGGCGGCGAATGTTCGTTACGTGCGTTTATAACGGACTACGTGCGCACAGGCGAAAGCGAGAGACTGGCTAGTATCGCGCGCGCCGCTATTGACGAGGCCATGAGATCTCCGTCCGCTTGGAGAGAAAGAGTTCCACCACCCGCTACCAACGCTAAATCTGGGGAGCGTCACCGCGTCGTGTTCGCGTGCTGCGCGTACAGCTGGCAGGCGGTGCACGGCGCGATGCGGCACGCGCGTCGCTGCGGCGCGAGCGGCGGCACGGAGGCGGCGCGCGCGGTGGCGGGTACATTGACCCCGCTGGCGACACAGTGCCGCGCGTGCCCGCACGCCGCGCCGCCCGCCGCCGCGCGCGCCGCCCGCTGGCTCGCCGACGACGACATCGTGCGGTTCCTGCAGTCGCTGCACAACTGGAAGGCGATGACCGGCGACGTGCCCGAAACCAAACAGAGCGCTCAGGAACTGAAACAGGCCTACGAACGGGAAGCCGAGATCTTGGGTTCGAGTTTGGGCGACGGCGCCGTTACACGCAAAGAAATCATATCGGACATGAATACGCTCGCCGACATAGCCGTCCTGTGCGAGACAATGGATTGGCTAAGTGCAAATATAAGAAGCATTCCCTCAATGTTATCCGGTGCAATAAAGCTCTCCGAGAAATTCCTGAACGACATATCTGCTGCAGCGTCTGTATTCGATGAGATAACACATAAATGCTTGCTTTTCTTGCATTTGGAGATGCGCATAGAGTGCTTTCATTATTTGGGTCAAGAGGATCGCGAAGGTGAAAACGGCTCCGAAGCGGACAGCCAACAGTCCGGAGGAGCAGCGCAGCTGGCCCACGCGCTGCTGGCCTTCCACGAACACGCGTCGGCTGTACTGGCGCCCTCTAGGCTCGCTTATATATTCAACGGTATAGGCGAGATGATGTCGGCCGCGGTAGTGTGGAGGTGGCAGGGGGAGCGCGGGGCGGAGGGCGGGGCGGCGGCTCGACTCGCGACACTGCGCCACTGTCTCGCTGCGCTACATCTACCTCACGATGGCTTACATAGAGCGCACGCGTACTTACATCTGTTAGCGTGTACTCCGCAGGAAATAATAACGTCAGTACGCGAAAAGGGTGCTCAATTCACGGAGTTGGAGTATTTGAACGCTTTCAAGGTTATCGGCGTTCGGCGTAACGTGTCCTCGGGCGACATGAAAATACATCTAAAACAACTATCGACCGCTCTGGGTCACGTCGGTGTTACCGTGTGA
Protein
MSSPPPTKPPRGVKQGKETSGLLMSVIRTLSASETNEQREKERAKLEKEYKKSDARLDELVAVHAQEITDVMQKFTAVGTALSVWGQHCEAAVARLGACRGLLRLRRDDLRRLWADARTHHYALQMLTDIERVVVSERETRESLSAGRVLAAAYTLKAALDTADTTLSHVDALNTTRQHLQAKKRELVDVIVRRVNDAVYRGETAPMTRRLSARRRTAILLAELTKNEEVTDAYLQDITPEYEASLSEDDKTFESTVMISLEALGVLEQLKEATEKFKVQIQNELLEVIDSVSRRIIDEGDVEIDQECDEDGETQEAEGVTEAGRPLARLVASLGEELRACAARNKRLLQLWRGALQKHRLSDATLHTEQHYWSAVQQVLQLLLTEYLEIESVNLAAQRSGTTTVEAAGSQLRLTDYFSKKRPQRQHTKLFRFTGPTAVTPSLQARRHGYPLVCRADPAHLHAVLPALHALCADIEPLTGGGECSLRAFITDYVRTGESERLASIARAAIDEAMRSPSAWRERVPPPATNAKSGERHRVVFACCAYSWQAVHGAMRHARRCGASGGTEAARAVAGTLTPLATQCRACPHAAPPAAARAARWLADDDIVRFLQSLHNWKAMTGDVPETKQSAQELKQAYEREAEILGSSLGDGAVTRKEIISDMNTLADIAVLCETMDWLSANIRSIPSMLSGAIKLSEKFLNDISAAASVFDEITHKCLLFLHLEMRIECFHYLGQEDREGENGSEADSQQSGGAAQLAHALLAFHEHASAVLAPSRLAYIFNGIGEMMSAAVVWRWQGERGAEGGAAARLATLRHCLAALHLPHDGLHRAHAYLHLLACTPQEIITSVREKGAQFTELEYLNAFKVIGVRRNVSSGDMKIHLKQLSTALGHVGVTV
Summary
Description
Component of the exocyst complex involved in the docking of exocytic vesicles with fusion sites on the plasma membrane. Involved in regulation of synaptic microtubule formation, and also regulation of synaptic growth and glutamate receptor trafficking. Does not appear to be required for basal neurotransmission.
Subunit
The exocyst complex is composed of Sec3/Exoc1, Sec5/Exoc2, Sec6/Exoc3, Sec8/Exoc4, Sec10/Exoc5, Sec15/Exoc6, exo70/Exoc7 and Exo84/Exoc8.
The exocyst complex is composed of EXOC1, EXOC2, EXOC3, EXOC4, EXOC5, EXOC6, EXOC7 and EXOC8. Interacts with BIRC6/bruce (By similarity). Interacts with MYRIP. Interacts with SH3BP1; required for the localization of both SH3BP1 and the exocyst to the leading edge of migrating cells (By similarity).
The exocyst complex is composed of EXOC1, EXOC2, EXOC3, EXOC4, EXOC5, EXOC6, EXOC7 and EXOC8 (By similarity). Interacts with BIRC6/bruce (By similarity). Interacts with MYRIP. Interacts with SH3BP1; required for the localization of both SH3BP1 and the exocyst to the leading edge of migrating cells (By similarity).
The exocyst complex is composed of EXOC1, EXOC2, EXOC3, EXOC4, EXOC5, EXOC6, EXOC7 and EXOC8. Interacts with BIRC6/bruce (By similarity). Interacts with MYRIP. Interacts with SH3BP1; required for the localization of both SH3BP1 and the exocyst to the leading edge of migrating cells (By similarity).
The exocyst complex is composed of EXOC1, EXOC2, EXOC3, EXOC4, EXOC5, EXOC6, EXOC7 and EXOC8 (By similarity). Interacts with BIRC6/bruce (By similarity). Interacts with MYRIP. Interacts with SH3BP1; required for the localization of both SH3BP1 and the exocyst to the leading edge of migrating cells (By similarity).
Similarity
Belongs to the SEC8 family.
Keywords
Coiled coil
Complete proteome
Exocytosis
Phosphoprotein
Protein transport
Reference proteome
Transport
Acetylation
Cell projection
Feature
chain Exocyst complex component 4
Uniprot
A0A2A4JLQ5
A0A2H1VE64
A0A3S2LH57
A0A1Y1MN89
A0A2J7RD24
A0A2R7W5Z1
+ More
E0VYL8 A0A1S4FCS9 A0A0A1WKB7 A0A034VXR8 N6UTL6 W8C7F2 A0A0A9WJT2 Q177C5 A0A0M5J293 N6UM97 A0A0P8XZU2 A0A0P8YNW5 A0A0P9CB80 A0A0P9ASV3 A0A0P8Y106 A0A0P9A174 U3PXA7 Q9VNH6 B4QXJ5 A0A0R1E4E3 A0A0R1E437 B4PUX9 A0A0R1E4M2 B0W348 B4I471 B4G494 B3P2J6 B4LYP8 A0A0Q9WPT0 A0A0Q9WEQ2 A0A0Q9WQM2 A0A0Q9WM61 B4JGG6 A0A0Q9WR03 A0A3B0KBU3 A0A182PSL1 A0A1Q3F4T8 A0A1Q3F4V2 A0A3B0JLP9 A0A0R3NHL8 A0A0R3NHU1 A0A0R3NHK5 A0A0R3NHP2 A0A0R3NHX4 A0A0R3NHJ2 A0A0R3NN99 Q297L0 A0A1W4UWY8 A0A1W4UWN3 A0A1W4UYC1 A0A1W4UJ65 A0A1W4UWY3 A0A182GVJ4 A0A1A9US47 A0A1B6JSP4 A0A1I8MP57 A0A1B0C0Z3 A0A0C9R726 A0A224XIS8 A0A1B6EHB1 B4NJI7 A0A1I8Q9N4 J9JN16 A0A1A9X8H5 A0A023F027 D6X531 A0A0T6BEF7 A0A1B6M4H7 A0A1L8GZQ8 Q52KN6 G1MEJ2 A0A2Y9T8J7 A0A341AT54 A0A383ZQ17 G3T7D1 M3X5B6 M0RBF8 A0A2K5EGU0 A0A2K6U1P2 Q62824 U3F757 Q69ZD1 O35382
E0VYL8 A0A1S4FCS9 A0A0A1WKB7 A0A034VXR8 N6UTL6 W8C7F2 A0A0A9WJT2 Q177C5 A0A0M5J293 N6UM97 A0A0P8XZU2 A0A0P8YNW5 A0A0P9CB80 A0A0P9ASV3 A0A0P8Y106 A0A0P9A174 U3PXA7 Q9VNH6 B4QXJ5 A0A0R1E4E3 A0A0R1E437 B4PUX9 A0A0R1E4M2 B0W348 B4I471 B4G494 B3P2J6 B4LYP8 A0A0Q9WPT0 A0A0Q9WEQ2 A0A0Q9WQM2 A0A0Q9WM61 B4JGG6 A0A0Q9WR03 A0A3B0KBU3 A0A182PSL1 A0A1Q3F4T8 A0A1Q3F4V2 A0A3B0JLP9 A0A0R3NHL8 A0A0R3NHU1 A0A0R3NHK5 A0A0R3NHP2 A0A0R3NHX4 A0A0R3NHJ2 A0A0R3NN99 Q297L0 A0A1W4UWY8 A0A1W4UWN3 A0A1W4UYC1 A0A1W4UJ65 A0A1W4UWY3 A0A182GVJ4 A0A1A9US47 A0A1B6JSP4 A0A1I8MP57 A0A1B0C0Z3 A0A0C9R726 A0A224XIS8 A0A1B6EHB1 B4NJI7 A0A1I8Q9N4 J9JN16 A0A1A9X8H5 A0A023F027 D6X531 A0A0T6BEF7 A0A1B6M4H7 A0A1L8GZQ8 Q52KN6 G1MEJ2 A0A2Y9T8J7 A0A341AT54 A0A383ZQ17 G3T7D1 M3X5B6 M0RBF8 A0A2K5EGU0 A0A2K6U1P2 Q62824 U3F757 Q69ZD1 O35382
Pubmed
28004739
20566863
17510324
25830018
25348373
23537049
+ More
24495485 25401762 26823975 17994087 18057021 16351720 10731132 12537572 12537569 18327897 17550304 15632085 23185243 26483478 25315136 25474469 18362917 19820115 27762356 20010809 17975172 15057822 22673903 7568183 17827149 21658605 25243066 15368895 9441674 15489334 21183079
24495485 25401762 26823975 17994087 18057021 16351720 10731132 12537572 12537569 18327897 17550304 15632085 23185243 26483478 25315136 25474469 18362917 19820115 27762356 20010809 17975172 15057822 22673903 7568183 17827149 21658605 25243066 15368895 9441674 15489334 21183079
EMBL
NWSH01001154
PCG72340.1
ODYU01001879
SOQ38702.1
RSAL01000125
RVE46593.1
+ More
GEZM01026226 JAV87132.1 NEVH01005303 PNF38735.1 KK854366 PTY15102.1 DS235845 EEB18474.1 GBXI01014788 JAC99503.1 GAKP01010841 JAC48111.1 APGK01016463 KB739858 ENN82092.1 GAMC01000587 JAC05969.1 GBHO01036831 GBHO01036828 GDHC01004978 JAG06773.1 JAG06776.1 JAQ13651.1 CH477378 EAT42267.1 CP012526 ALC46761.1 APGK01017822 KB740053 KB632360 ENN81811.1 ERL93516.1 CH902617 KPU80453.1 KPU80454.1 KPU80455.1 KPU80450.1 KPU80451.1 KPU80452.1 KPU80456.1 BT150369 AGW52179.1 AY905551 AE014297 AY119660 CM000364 EDX11781.1 CM000160 KRK03961.1 KRK03964.1 EDW97786.2 KRK03962.1 KRK03965.1 KRK03967.1 DS231830 EDS30756.1 CH480821 EDW55014.1 CH479179 EDW25132.1 CH954181 EDV47946.1 CH940650 EDW66975.2 KRF83039.1 KRF83032.1 KRF83038.1 KRF83031.1 KRF83033.1 KRF83034.1 CH916369 EDV93663.1 KRF83035.1 OUUW01000007 SPP83156.1 GFDL01012475 JAV22570.1 GFDL01012453 JAV22592.1 SPP83155.1 CM000070 KRT00545.1 KRT00548.1 KRT00553.1 KRT00551.1 KRT00555.1 KRT00546.1 KRT00544.1 KRT00547.1 EAL28195.2 JXUM01019055 JXUM01019056 KQ560531 KXJ81967.1 GECU01005497 JAT02210.1 JXJN01023829 GBYB01002566 JAG72333.1 GFTR01007934 JAW08492.1 GEDC01000034 JAS37264.1 CH964272 EDW83911.1 ABLF02040311 GBBI01004321 JAC14391.1 KQ971382 EEZ97195.1 LJIG01001216 KRT85724.1 GEBQ01009146 JAT30831.1 CM004470 OCT89261.1 BC094262 AAH94262.1 ACTA01107034 ACTA01115034 ACTA01123034 ACTA01131034 ACTA01139034 ACTA01147033 ACTA01155032 ACTA01163031 ACTA01171029 ACTA01179027 AANG04000096 AABR07060165 AABR07060166 AABR07060167 AABR07060168 AABR07060169 AABR07060170 AABR07060171 AABR07060172 AABR07060173 AABR07060174 U32498 GAMT01006800 GAMS01007446 GAMR01008084 GAMQ01007070 GAMP01000616 JAB05061.1 JAB15690.1 JAB25848.1 JAB34781.1 JAB52139.1 AK173235 BAD32513.1 AF022962 BC034644
GEZM01026226 JAV87132.1 NEVH01005303 PNF38735.1 KK854366 PTY15102.1 DS235845 EEB18474.1 GBXI01014788 JAC99503.1 GAKP01010841 JAC48111.1 APGK01016463 KB739858 ENN82092.1 GAMC01000587 JAC05969.1 GBHO01036831 GBHO01036828 GDHC01004978 JAG06773.1 JAG06776.1 JAQ13651.1 CH477378 EAT42267.1 CP012526 ALC46761.1 APGK01017822 KB740053 KB632360 ENN81811.1 ERL93516.1 CH902617 KPU80453.1 KPU80454.1 KPU80455.1 KPU80450.1 KPU80451.1 KPU80452.1 KPU80456.1 BT150369 AGW52179.1 AY905551 AE014297 AY119660 CM000364 EDX11781.1 CM000160 KRK03961.1 KRK03964.1 EDW97786.2 KRK03962.1 KRK03965.1 KRK03967.1 DS231830 EDS30756.1 CH480821 EDW55014.1 CH479179 EDW25132.1 CH954181 EDV47946.1 CH940650 EDW66975.2 KRF83039.1 KRF83032.1 KRF83038.1 KRF83031.1 KRF83033.1 KRF83034.1 CH916369 EDV93663.1 KRF83035.1 OUUW01000007 SPP83156.1 GFDL01012475 JAV22570.1 GFDL01012453 JAV22592.1 SPP83155.1 CM000070 KRT00545.1 KRT00548.1 KRT00553.1 KRT00551.1 KRT00555.1 KRT00546.1 KRT00544.1 KRT00547.1 EAL28195.2 JXUM01019055 JXUM01019056 KQ560531 KXJ81967.1 GECU01005497 JAT02210.1 JXJN01023829 GBYB01002566 JAG72333.1 GFTR01007934 JAW08492.1 GEDC01000034 JAS37264.1 CH964272 EDW83911.1 ABLF02040311 GBBI01004321 JAC14391.1 KQ971382 EEZ97195.1 LJIG01001216 KRT85724.1 GEBQ01009146 JAT30831.1 CM004470 OCT89261.1 BC094262 AAH94262.1 ACTA01107034 ACTA01115034 ACTA01123034 ACTA01131034 ACTA01139034 ACTA01147033 ACTA01155032 ACTA01163031 ACTA01171029 ACTA01179027 AANG04000096 AABR07060165 AABR07060166 AABR07060167 AABR07060168 AABR07060169 AABR07060170 AABR07060171 AABR07060172 AABR07060173 AABR07060174 U32498 GAMT01006800 GAMS01007446 GAMR01008084 GAMQ01007070 GAMP01000616 JAB05061.1 JAB15690.1 JAB25848.1 JAB34781.1 JAB52139.1 AK173235 BAD32513.1 AF022962 BC034644
Proteomes
UP000218220
UP000283053
UP000235965
UP000009046
UP000019118
UP000008820
+ More
UP000092553 UP000030742 UP000007801 UP000000803 UP000000304 UP000002282 UP000002320 UP000001292 UP000008744 UP000008711 UP000008792 UP000001070 UP000268350 UP000075885 UP000001819 UP000192221 UP000069940 UP000249989 UP000078200 UP000095301 UP000092460 UP000007798 UP000095300 UP000007819 UP000092443 UP000007266 UP000186698 UP000008912 UP000248484 UP000252040 UP000261681 UP000007646 UP000011712 UP000002494 UP000233020 UP000233220 UP000008225 UP000000589
UP000092553 UP000030742 UP000007801 UP000000803 UP000000304 UP000002282 UP000002320 UP000001292 UP000008744 UP000008711 UP000008792 UP000001070 UP000268350 UP000075885 UP000001819 UP000192221 UP000069940 UP000249989 UP000078200 UP000095301 UP000092460 UP000007798 UP000095300 UP000007819 UP000092443 UP000007266 UP000186698 UP000008912 UP000248484 UP000252040 UP000261681 UP000007646 UP000011712 UP000002494 UP000233020 UP000233220 UP000008225 UP000000589
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A2A4JLQ5
A0A2H1VE64
A0A3S2LH57
A0A1Y1MN89
A0A2J7RD24
A0A2R7W5Z1
+ More
E0VYL8 A0A1S4FCS9 A0A0A1WKB7 A0A034VXR8 N6UTL6 W8C7F2 A0A0A9WJT2 Q177C5 A0A0M5J293 N6UM97 A0A0P8XZU2 A0A0P8YNW5 A0A0P9CB80 A0A0P9ASV3 A0A0P8Y106 A0A0P9A174 U3PXA7 Q9VNH6 B4QXJ5 A0A0R1E4E3 A0A0R1E437 B4PUX9 A0A0R1E4M2 B0W348 B4I471 B4G494 B3P2J6 B4LYP8 A0A0Q9WPT0 A0A0Q9WEQ2 A0A0Q9WQM2 A0A0Q9WM61 B4JGG6 A0A0Q9WR03 A0A3B0KBU3 A0A182PSL1 A0A1Q3F4T8 A0A1Q3F4V2 A0A3B0JLP9 A0A0R3NHL8 A0A0R3NHU1 A0A0R3NHK5 A0A0R3NHP2 A0A0R3NHX4 A0A0R3NHJ2 A0A0R3NN99 Q297L0 A0A1W4UWY8 A0A1W4UWN3 A0A1W4UYC1 A0A1W4UJ65 A0A1W4UWY3 A0A182GVJ4 A0A1A9US47 A0A1B6JSP4 A0A1I8MP57 A0A1B0C0Z3 A0A0C9R726 A0A224XIS8 A0A1B6EHB1 B4NJI7 A0A1I8Q9N4 J9JN16 A0A1A9X8H5 A0A023F027 D6X531 A0A0T6BEF7 A0A1B6M4H7 A0A1L8GZQ8 Q52KN6 G1MEJ2 A0A2Y9T8J7 A0A341AT54 A0A383ZQ17 G3T7D1 M3X5B6 M0RBF8 A0A2K5EGU0 A0A2K6U1P2 Q62824 U3F757 Q69ZD1 O35382
E0VYL8 A0A1S4FCS9 A0A0A1WKB7 A0A034VXR8 N6UTL6 W8C7F2 A0A0A9WJT2 Q177C5 A0A0M5J293 N6UM97 A0A0P8XZU2 A0A0P8YNW5 A0A0P9CB80 A0A0P9ASV3 A0A0P8Y106 A0A0P9A174 U3PXA7 Q9VNH6 B4QXJ5 A0A0R1E4E3 A0A0R1E437 B4PUX9 A0A0R1E4M2 B0W348 B4I471 B4G494 B3P2J6 B4LYP8 A0A0Q9WPT0 A0A0Q9WEQ2 A0A0Q9WQM2 A0A0Q9WM61 B4JGG6 A0A0Q9WR03 A0A3B0KBU3 A0A182PSL1 A0A1Q3F4T8 A0A1Q3F4V2 A0A3B0JLP9 A0A0R3NHL8 A0A0R3NHU1 A0A0R3NHK5 A0A0R3NHP2 A0A0R3NHX4 A0A0R3NHJ2 A0A0R3NN99 Q297L0 A0A1W4UWY8 A0A1W4UWN3 A0A1W4UYC1 A0A1W4UJ65 A0A1W4UWY3 A0A182GVJ4 A0A1A9US47 A0A1B6JSP4 A0A1I8MP57 A0A1B0C0Z3 A0A0C9R726 A0A224XIS8 A0A1B6EHB1 B4NJI7 A0A1I8Q9N4 J9JN16 A0A1A9X8H5 A0A023F027 D6X531 A0A0T6BEF7 A0A1B6M4H7 A0A1L8GZQ8 Q52KN6 G1MEJ2 A0A2Y9T8J7 A0A341AT54 A0A383ZQ17 G3T7D1 M3X5B6 M0RBF8 A0A2K5EGU0 A0A2K6U1P2 Q62824 U3F757 Q69ZD1 O35382
Ontologies
GO
GO:0000145
GO:0090522
GO:0006904
GO:0016028
GO:0000212
GO:0009925
GO:0007009
GO:0007298
GO:0016324
GO:0007286
GO:0007110
GO:0007111
GO:0045887
GO:0000916
GO:0007269
GO:0016080
GO:0050803
GO:0016192
GO:0005737
GO:0016081
GO:0098793
GO:0016020
GO:0015031
GO:0005886
GO:0016021
GO:0032584
GO:0006612
GO:0045202
GO:0007268
GO:0006887
GO:0006893
GO:0047485
GO:0017160
GO:0035748
GO:0048341
GO:0030165
GO:0005902
GO:0044091
GO:0050850
GO:0043025
GO:0032991
GO:0031252
GO:0043209
GO:0006903
GO:0090543
GO:0030426
GO:0048709
GO:0030010
GO:0043005
GO:0055108
GO:0043198
GO:0051223
GO:0014069
GO:0008021
GO:0044877
PANTHER
Topology
Subcellular location
Midbody
Colocalizes with CNTRL/centriolin at the midbody ring (By similarity). Localizes at the leading edge of migrating cells (PubMed:21658605). With evidence from 3 publications.
Midbody ring Colocalizes with CNTRL/centriolin at the midbody ring (By similarity). Localizes at the leading edge of migrating cells (PubMed:21658605). With evidence from 3 publications.
Cell projection Colocalizes with CNTRL/centriolin at the midbody ring (By similarity). Localizes at the leading edge of migrating cells (PubMed:21658605). With evidence from 3 publications.
Midbody ring Colocalizes with CNTRL/centriolin at the midbody ring (By similarity). Localizes at the leading edge of migrating cells (PubMed:21658605). With evidence from 3 publications.
Cell projection Colocalizes with CNTRL/centriolin at the midbody ring (By similarity). Localizes at the leading edge of migrating cells (PubMed:21658605). With evidence from 3 publications.
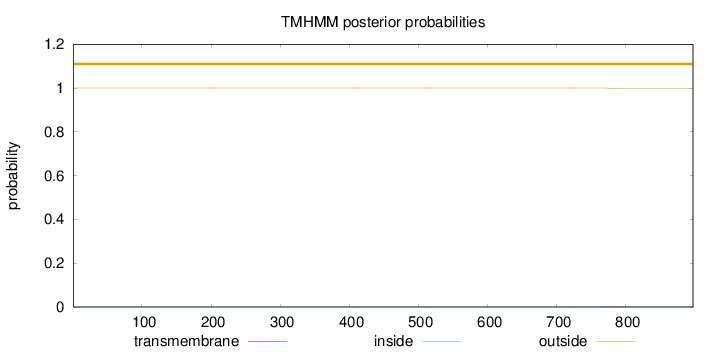
Length:
897
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00005
outside
1 - 897
Population Genetic Test Statistics
Pi
296.445444
Theta
172.42627
Tajima's D
2.163553
CLR
0.359698
CSRT
0.908204589770511
Interpretation
Uncertain
