Gene
KWMTBOMO13332
Pre Gene Modal
BGIBMGA010911
Annotation
PREDICTED:_rap1_GTPase-GDP_dissociation_stimulator_1-B_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.973 Golgi Reliability : 1.276
Sequence
CDS
ATGGATGAATGTTCAGCAACGAAATCTACTTCCTTAGAAACGTTGGTTATTCAAAATATAAGTAATGTGAAAGAACTAAAGTCGAAATTAAGTGAAATAATTAAAAAAGGAAAAGACTATGAATTTGATGTTTCTTCGTGCGTCAAGACTTTAATAAAGAGCAACGACCAAGAGATAGTATTCCTGACTGTCGAAGCCATATCAGAGCTCGTTAAATGCGACGAGAAACGTCGAACTTATTCTCAGAAAGAGATAATCAGCCCCATTCTAGATATTCTGCAAAAAGAAATAACTACCGAAAATGCTGCGTTAATAAAACAGTGCTTTCGAGCTCTCGGTAACTTGTGCTGTGACTGTGACTCGTCAAGAAAGATAATCTTGGAATGTGGAGGTGTTCCCAACATAATAAATCTAATGAAAAAATATTTCAGTGCCCACAGTCAATTAAGCATGTATGCATTTAAAACTTTATTAAATTTTGCGATCGGTGGTCAAGAATTCACAAATGCAATGGTTGATGGTGGAGTAGTTGAAATGCTTCAAATGGTTGTCATTCAGGAGCTAGAGAAAGAGGAAATTGATGAAGAGGCTGTTTGTGCTTTGCTTTCCTTATTATCTCTTATCAACGAAAATGATACAGAAATATTGTACAGTGATGAAATTAATGCAGGGGTTTTGAAAGTCCTAACTAATACTGCTGATATTGATGTTTCGGAGTTATGCCTCGATATACTTCATGCCCAAGCTGAGCATGATTCTGTAAAGACCTTAATAGCAAAACATGGTGGTATCCAAGTAGTTTGTTCTCGTATTGAACAGCTAATGCAAAAGTTTGATCACGGAGAACTTGATGCAGATCAATCTGATGTTGAGGCCATCCTGAAGCAAGCTTGTGATTTAATAGTCATTGTTTTGACTGGAGATGAAGCAATGCACATACTATACAACAAAGGTCTCGGTGAAGTTTACATCTCAATGGTAAAATGGCTGGAGCACACTAACTACCACCTAATAACTACAGGTGTCCTTGCTATTGGAAACTTTGCCAGACAAGATGACTATTGTGCACAGATGATGGACGATAAGATATATGATAAATTGTTGGATCTCTTTGATGTATATGTAAGTTTCGGAGTTCAAATGCAAAAGGAACATAGTGTGTTACATCCGATAGAACCTGGAACCATCACAAAGATCCAGCATGCTATATTGTCCGCTCTCCGGAACCTCACGGTGCCTGTGGCGAATAAAAGGATAATAGCGGCTCAGGGAAGGGCTGCGCCTACGTTGCTGGGGGCATTGCTTTCGGTAGAAGATCATCATGTAGCTTACAAACTTCTGGCTGCCATCAGGATGCTTGTTGATGGTCAAGAGAGTGTTGCCCGTCTGTTGGGCCGTCACAAGTCAGCGTTGCGTTGCGTGGCGCGCTGGGGGCACGCGGGGGAGTACGCGGGCGCGGCGGGGGAGGCGCCCCGCCTGCTCGCCTGGGCCGTCAAGCTTCTCACGCGCGGACCCTACTGGCACAATATCGTCGAGGTGGACGGCTGTATATCCAGTTTAGTCAATATGCTGGTGGCCAGTCACTCGCTAATGCAGAACGAAGCCATTCTCGCTCTGACGCTATTAGCTATTGAAACATTAAAAAATATTTCTCCCTCGAGTGAACATCCAGAATTCGATTACGAAAAAAGCTTTATCTCTCAGTTGATTAAGTCGGAAATCGGTAAGCACGTTTCCATACTAGTCGATACGAACTGCGCTAAAATGCCCGTCGAGGTTGCGGAGAACCTTCTCGCCTTCCTAGACATCACTTCGAAGAAGAATAAATTAGCTCTCGATTATAAGGAGGCTAAAGTGACAGAGTCCTTGAAGAAGTTCGCTGAATCCAGACAGGATCTAAGCGAGGATCTCAAAAATTGTATCACTGGTGTAATCGCAGCTATCTCAGACAATGGGAAAGATTAA
Protein
MDECSATKSTSLETLVIQNISNVKELKSKLSEIIKKGKDYEFDVSSCVKTLIKSNDQEIVFLTVEAISELVKCDEKRRTYSQKEIISPILDILQKEITTENAALIKQCFRALGNLCCDCDSSRKIILECGGVPNIINLMKKYFSAHSQLSMYAFKTLLNFAIGGQEFTNAMVDGGVVEMLQMVVIQELEKEEIDEEAVCALLSLLSLINENDTEILYSDEINAGVLKVLTNTADIDVSELCLDILHAQAEHDSVKTLIAKHGGIQVVCSRIEQLMQKFDHGELDADQSDVEAILKQACDLIVIVLTGDEAMHILYNKGLGEVYISMVKWLEHTNYHLITTGVLAIGNFARQDDYCAQMMDDKIYDKLLDLFDVYVSFGVQMQKEHSVLHPIEPGTITKIQHAILSALRNLTVPVANKRIIAAQGRAAPTLLGALLSVEDHHVAYKLLAAIRMLVDGQESVARLLGRHKSALRCVARWGHAGEYAGAAGEAPRLLAWAVKLLTRGPYWHNIVEVDGCISSLVNMLVASHSLMQNEAILALTLLAIETLKNISPSSEHPEFDYEKSFISQLIKSEIGKHVSILVDTNCAKMPVEVAENLLAFLDITSKKNKLALDYKEAKVTESLKKFAESRQDLSEDLKNCITGVIAAISDNGKD
Summary
Uniprot
H9JN08
A0A2A4JLP4
A0A3S2LXN4
A0A2H1VCX0
A0A212F5R3
A0A0L7L8E8
+ More
A0A194Q414 D6WH32 A0A1Y1NF21 A0A0K8WDW2 W8AYZ1 A0A034W070 A0A0A1WZM9 A0A1I8P7R5 A0A0T6AZM0 A0A1I8MLE0 A0A1L8DMJ5 A0A067QY97 B4MPU6 A0A1B0CRW1 A0A0L0CAM7 A0A1A9WI19 B3MGR8 B5E1D1 A0A0J9R734 B4GCC0 B4HQE6 A0A0M4EFY2 B4J3V7 A0A1W4XSG1 A1Z6S6 A1Z6S7 O44116 A0A1W4VEZ9 B3NAK2 B4MFA9 Q86LF4 B4P1B4 A0A3B0JIL2 B4KU52 F4WJA3 A0A182FEC6 A0A2M4BI12 W5JQZ2 A0A158NEB4 A0A182LUM6 A0A2M4AM01 A0A182RP34 A0A0J7NEZ5 A0A2M4BHD2 A0A3L8DBB6 A0A2M3ZID7 A0A026W5V3 A0A182YBY2 A0A182PQ88 A0A1B6JE39 A0A182UH60 Q7Q9E6 E2A0C9 A0A182HNS2 A0A182HDP9 A0A182X8G9 A0A1Q3F8P2 A0A195BX53 Q17LP7 A0A151WT72 A0A182WMH7 A0A1S4EY80 A0A195FL32 E9J1I1 A0A1B6GG94 A0A182VKH8 A0A182T3B6 A0A182JHJ3 A0A2P8Y1J1 Q8T996 A0A182KGH2 E2C9A2 A0A0C9RCJ9 A0A182QMR3 A0A084W9M5 A0A195CCD2 A0A151J6H7 A0A0A9YZP0 B0W5K1 A0A023EX93 A0A182NV12 B4QDT4 A0A023EWZ0 A0A1J1JAD5 A0A224XLG5 A0A2J7Q7K4 A0A154NY23 A0A0L8HR07
A0A194Q414 D6WH32 A0A1Y1NF21 A0A0K8WDW2 W8AYZ1 A0A034W070 A0A0A1WZM9 A0A1I8P7R5 A0A0T6AZM0 A0A1I8MLE0 A0A1L8DMJ5 A0A067QY97 B4MPU6 A0A1B0CRW1 A0A0L0CAM7 A0A1A9WI19 B3MGR8 B5E1D1 A0A0J9R734 B4GCC0 B4HQE6 A0A0M4EFY2 B4J3V7 A0A1W4XSG1 A1Z6S6 A1Z6S7 O44116 A0A1W4VEZ9 B3NAK2 B4MFA9 Q86LF4 B4P1B4 A0A3B0JIL2 B4KU52 F4WJA3 A0A182FEC6 A0A2M4BI12 W5JQZ2 A0A158NEB4 A0A182LUM6 A0A2M4AM01 A0A182RP34 A0A0J7NEZ5 A0A2M4BHD2 A0A3L8DBB6 A0A2M3ZID7 A0A026W5V3 A0A182YBY2 A0A182PQ88 A0A1B6JE39 A0A182UH60 Q7Q9E6 E2A0C9 A0A182HNS2 A0A182HDP9 A0A182X8G9 A0A1Q3F8P2 A0A195BX53 Q17LP7 A0A151WT72 A0A182WMH7 A0A1S4EY80 A0A195FL32 E9J1I1 A0A1B6GG94 A0A182VKH8 A0A182T3B6 A0A182JHJ3 A0A2P8Y1J1 Q8T996 A0A182KGH2 E2C9A2 A0A0C9RCJ9 A0A182QMR3 A0A084W9M5 A0A195CCD2 A0A151J6H7 A0A0A9YZP0 B0W5K1 A0A023EX93 A0A182NV12 B4QDT4 A0A023EWZ0 A0A1J1JAD5 A0A224XLG5 A0A2J7Q7K4 A0A154NY23 A0A0L8HR07
Pubmed
19121390
22118469
26227816
26354079
18362917
19820115
+ More
28004739 24495485 25348373 25830018 25315136 24845553 17994087 26108605 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9533953 17550304 21719571 20920257 23761445 21347285 30249741 24508170 25244985 12364791 20798317 26483478 17510324 21282665 29403074 24438588 25401762 26823975 24945155 25474469
28004739 24495485 25348373 25830018 25315136 24845553 17994087 26108605 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9533953 17550304 21719571 20920257 23761445 21347285 30249741 24508170 25244985 12364791 20798317 26483478 17510324 21282665 29403074 24438588 25401762 26823975 24945155 25474469
EMBL
BABH01030648
NWSH01001154
PCG72332.1
RSAL01000125
RVE46591.1
ODYU01001879
+ More
SOQ38700.1 AGBW02010144 OWR49077.1 JTDY01002353 KOB71604.1 KQ459586 KPI98150.1 KQ971321 EFA00118.1 GEZM01003833 JAV96533.1 GDHF01024590 GDHF01003259 JAI27724.1 JAI49055.1 GAMC01012545 JAB94010.1 GAKP01010868 GAKP01010865 JAC48084.1 GBXI01010171 GBXI01003297 JAD04121.1 JAD10995.1 LJIG01022488 KRT80317.1 GFDF01006453 JAV07631.1 KK853114 KDR11195.1 CH963849 EDW74135.1 AJWK01025352 AJWK01025353 JRES01000678 KNC29290.1 CH902619 EDV36826.2 CM000071 EDY68736.2 CM002911 KMY91886.1 CH479181 EDW31438.1 CH480816 EDW46685.1 CP012524 ALC42351.1 CH916367 EDW01540.1 AE013599 BT125966 AAF57418.1 ADX35945.1 BT044558 AAS64794.1 ACI15753.1 AF034421 AAB87984.1 CH954177 EDV59756.2 CH940667 EDW57278.2 BT004891 AAO47869.1 CM000157 EDW89116.2 OUUW01000001 SPP73066.1 CH933808 EDW08629.2 GL888182 EGI65681.1 GGFJ01003307 MBW52448.1 ADMH02000689 ETN65320.1 ADTU01013237 AXCM01011633 GGFK01008488 MBW41809.1 LBMM01005943 KMQ91075.1 GGFJ01003306 MBW52447.1 QOIP01000010 RLU17626.1 GGFM01007555 MBW28306.1 KK107455 EZA50414.1 GECU01010271 JAS97435.1 AAAB01008900 EAA09487.3 GL435569 EFN73132.1 APCN01002756 JXUM01034642 KQ561031 KXJ79962.1 GFDL01011129 JAV23916.1 KQ976401 KYM92543.1 CH477213 EAT47655.1 KQ982769 KYQ50825.1 KQ981490 KYN41056.1 GL767671 EFZ13316.1 GECZ01008307 JAS61462.1 PYGN01001050 PSN38110.1 AY069865 AAL40010.1 GL453806 EFN75447.1 GBYB01004646 JAG74413.1 AXCN02001010 ATLV01021841 KE525324 KFB46919.1 KQ978023 KYM97891.1 KQ979851 KYN18827.1 GBHO01005107 GBHO01005105 GDHC01006613 JAG38497.1 JAG38499.1 JAQ12016.1 DS231843 EDS35484.1 GAPW01000589 JAC13009.1 CM000362 EDX05946.1 GBBI01005009 JAC13703.1 CVRI01000075 CRL08518.1 GFTR01007156 JAW09270.1 NEVH01017442 PNF24564.1 KQ434781 KZC04481.1 KQ417497 KOF91656.1
SOQ38700.1 AGBW02010144 OWR49077.1 JTDY01002353 KOB71604.1 KQ459586 KPI98150.1 KQ971321 EFA00118.1 GEZM01003833 JAV96533.1 GDHF01024590 GDHF01003259 JAI27724.1 JAI49055.1 GAMC01012545 JAB94010.1 GAKP01010868 GAKP01010865 JAC48084.1 GBXI01010171 GBXI01003297 JAD04121.1 JAD10995.1 LJIG01022488 KRT80317.1 GFDF01006453 JAV07631.1 KK853114 KDR11195.1 CH963849 EDW74135.1 AJWK01025352 AJWK01025353 JRES01000678 KNC29290.1 CH902619 EDV36826.2 CM000071 EDY68736.2 CM002911 KMY91886.1 CH479181 EDW31438.1 CH480816 EDW46685.1 CP012524 ALC42351.1 CH916367 EDW01540.1 AE013599 BT125966 AAF57418.1 ADX35945.1 BT044558 AAS64794.1 ACI15753.1 AF034421 AAB87984.1 CH954177 EDV59756.2 CH940667 EDW57278.2 BT004891 AAO47869.1 CM000157 EDW89116.2 OUUW01000001 SPP73066.1 CH933808 EDW08629.2 GL888182 EGI65681.1 GGFJ01003307 MBW52448.1 ADMH02000689 ETN65320.1 ADTU01013237 AXCM01011633 GGFK01008488 MBW41809.1 LBMM01005943 KMQ91075.1 GGFJ01003306 MBW52447.1 QOIP01000010 RLU17626.1 GGFM01007555 MBW28306.1 KK107455 EZA50414.1 GECU01010271 JAS97435.1 AAAB01008900 EAA09487.3 GL435569 EFN73132.1 APCN01002756 JXUM01034642 KQ561031 KXJ79962.1 GFDL01011129 JAV23916.1 KQ976401 KYM92543.1 CH477213 EAT47655.1 KQ982769 KYQ50825.1 KQ981490 KYN41056.1 GL767671 EFZ13316.1 GECZ01008307 JAS61462.1 PYGN01001050 PSN38110.1 AY069865 AAL40010.1 GL453806 EFN75447.1 GBYB01004646 JAG74413.1 AXCN02001010 ATLV01021841 KE525324 KFB46919.1 KQ978023 KYM97891.1 KQ979851 KYN18827.1 GBHO01005107 GBHO01005105 GDHC01006613 JAG38497.1 JAG38499.1 JAQ12016.1 DS231843 EDS35484.1 GAPW01000589 JAC13009.1 CM000362 EDX05946.1 GBBI01005009 JAC13703.1 CVRI01000075 CRL08518.1 GFTR01007156 JAW09270.1 NEVH01017442 PNF24564.1 KQ434781 KZC04481.1 KQ417497 KOF91656.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000037510
UP000053268
+ More
UP000007266 UP000095300 UP000095301 UP000027135 UP000007798 UP000092461 UP000037069 UP000091820 UP000007801 UP000001819 UP000008744 UP000001292 UP000092553 UP000001070 UP000192223 UP000000803 UP000192221 UP000008711 UP000008792 UP000002282 UP000268350 UP000009192 UP000007755 UP000069272 UP000000673 UP000005205 UP000075883 UP000075900 UP000036403 UP000279307 UP000053097 UP000076408 UP000075885 UP000075902 UP000007062 UP000000311 UP000075840 UP000069940 UP000249989 UP000076407 UP000078540 UP000008820 UP000075809 UP000075920 UP000078541 UP000075903 UP000075901 UP000075880 UP000245037 UP000075881 UP000008237 UP000075886 UP000030765 UP000078542 UP000078492 UP000002320 UP000075884 UP000000304 UP000183832 UP000235965 UP000076502 UP000053454
UP000007266 UP000095300 UP000095301 UP000027135 UP000007798 UP000092461 UP000037069 UP000091820 UP000007801 UP000001819 UP000008744 UP000001292 UP000092553 UP000001070 UP000192223 UP000000803 UP000192221 UP000008711 UP000008792 UP000002282 UP000268350 UP000009192 UP000007755 UP000069272 UP000000673 UP000005205 UP000075883 UP000075900 UP000036403 UP000279307 UP000053097 UP000076408 UP000075885 UP000075902 UP000007062 UP000000311 UP000075840 UP000069940 UP000249989 UP000076407 UP000078540 UP000008820 UP000075809 UP000075920 UP000078541 UP000075903 UP000075901 UP000075880 UP000245037 UP000075881 UP000008237 UP000075886 UP000030765 UP000078542 UP000078492 UP000002320 UP000075884 UP000000304 UP000183832 UP000235965 UP000076502 UP000053454
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JN08
A0A2A4JLP4
A0A3S2LXN4
A0A2H1VCX0
A0A212F5R3
A0A0L7L8E8
+ More
A0A194Q414 D6WH32 A0A1Y1NF21 A0A0K8WDW2 W8AYZ1 A0A034W070 A0A0A1WZM9 A0A1I8P7R5 A0A0T6AZM0 A0A1I8MLE0 A0A1L8DMJ5 A0A067QY97 B4MPU6 A0A1B0CRW1 A0A0L0CAM7 A0A1A9WI19 B3MGR8 B5E1D1 A0A0J9R734 B4GCC0 B4HQE6 A0A0M4EFY2 B4J3V7 A0A1W4XSG1 A1Z6S6 A1Z6S7 O44116 A0A1W4VEZ9 B3NAK2 B4MFA9 Q86LF4 B4P1B4 A0A3B0JIL2 B4KU52 F4WJA3 A0A182FEC6 A0A2M4BI12 W5JQZ2 A0A158NEB4 A0A182LUM6 A0A2M4AM01 A0A182RP34 A0A0J7NEZ5 A0A2M4BHD2 A0A3L8DBB6 A0A2M3ZID7 A0A026W5V3 A0A182YBY2 A0A182PQ88 A0A1B6JE39 A0A182UH60 Q7Q9E6 E2A0C9 A0A182HNS2 A0A182HDP9 A0A182X8G9 A0A1Q3F8P2 A0A195BX53 Q17LP7 A0A151WT72 A0A182WMH7 A0A1S4EY80 A0A195FL32 E9J1I1 A0A1B6GG94 A0A182VKH8 A0A182T3B6 A0A182JHJ3 A0A2P8Y1J1 Q8T996 A0A182KGH2 E2C9A2 A0A0C9RCJ9 A0A182QMR3 A0A084W9M5 A0A195CCD2 A0A151J6H7 A0A0A9YZP0 B0W5K1 A0A023EX93 A0A182NV12 B4QDT4 A0A023EWZ0 A0A1J1JAD5 A0A224XLG5 A0A2J7Q7K4 A0A154NY23 A0A0L8HR07
A0A194Q414 D6WH32 A0A1Y1NF21 A0A0K8WDW2 W8AYZ1 A0A034W070 A0A0A1WZM9 A0A1I8P7R5 A0A0T6AZM0 A0A1I8MLE0 A0A1L8DMJ5 A0A067QY97 B4MPU6 A0A1B0CRW1 A0A0L0CAM7 A0A1A9WI19 B3MGR8 B5E1D1 A0A0J9R734 B4GCC0 B4HQE6 A0A0M4EFY2 B4J3V7 A0A1W4XSG1 A1Z6S6 A1Z6S7 O44116 A0A1W4VEZ9 B3NAK2 B4MFA9 Q86LF4 B4P1B4 A0A3B0JIL2 B4KU52 F4WJA3 A0A182FEC6 A0A2M4BI12 W5JQZ2 A0A158NEB4 A0A182LUM6 A0A2M4AM01 A0A182RP34 A0A0J7NEZ5 A0A2M4BHD2 A0A3L8DBB6 A0A2M3ZID7 A0A026W5V3 A0A182YBY2 A0A182PQ88 A0A1B6JE39 A0A182UH60 Q7Q9E6 E2A0C9 A0A182HNS2 A0A182HDP9 A0A182X8G9 A0A1Q3F8P2 A0A195BX53 Q17LP7 A0A151WT72 A0A182WMH7 A0A1S4EY80 A0A195FL32 E9J1I1 A0A1B6GG94 A0A182VKH8 A0A182T3B6 A0A182JHJ3 A0A2P8Y1J1 Q8T996 A0A182KGH2 E2C9A2 A0A0C9RCJ9 A0A182QMR3 A0A084W9M5 A0A195CCD2 A0A151J6H7 A0A0A9YZP0 B0W5K1 A0A023EX93 A0A182NV12 B4QDT4 A0A023EWZ0 A0A1J1JAD5 A0A224XLG5 A0A2J7Q7K4 A0A154NY23 A0A0L8HR07
PDB
5XGC
E-value=3.89789e-35,
Score=373
Ontologies
GO
PANTHER
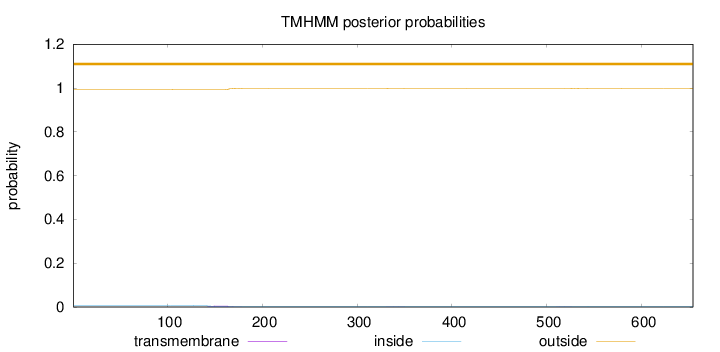
Topology
Length:
654
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14274
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00724
outside
1 - 654
Population Genetic Test Statistics
Pi
176.965537
Theta
186.487362
Tajima's D
-0.532261
CLR
1.843668
CSRT
0.24108794560272
Interpretation
Uncertain
