Gene
KWMTBOMO13331
Annotation
PREDICTED:_uncharacterized_protein_LOC105841419_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.643
Sequence
CDS
ATGGAATCAGTAACAACGAGAGTGAATTTGTTGGGCAACCCGACTAACCATCGGCGCCACCCAGTATCCGGGCTGGATGGTGTGAAATTGGCAACCGAAGCTTCCCCTGAGGTAAATCGACAGGCCTCAGAAGGAGGAGATACTACTGTCGGTACTGCAGCTTCTAAAAGCGAGAAGACCTCTATACGATCGAGTCATCGTATTGGCGCTTGGAACGTCCGAGGTCTGCTGGAGGCGGGTAAGCTGACAATCGTGGAAAAGGAAATGGAATCTCATGACATCTCCATACTAGGCATCAGTGAAACTCACATGCGTGGTAATGGCCACTTCACCACTGCGACAGGGAACACCATGTACTTCTCAGGCTGTGAAGATTCAAGTTTGAGTGGAGTCGGAATCATATTGCCGCCACACTTAAACAAATATGTCCTCGGTTATAATCCGGTGTCTGATAGAATCCTGACAATGAAACTGAATACCAATCCATGTGTGCTGAATGTTATACAGATATACGCACCGACGGCACAAGCTCAGGATGAGTTAATTGTGGACTTCTATGGAAAACTGGAGGAAATACTGAATTCTATACCTAGTCGGGAAATTAAGATAATTCTTGGTGATTGGAATGCCAAAGTTGGCAGTACACTGGATGATAATCATATCAGGAGCACAGTTGGAAAATTTGGACTTGGCATTAGGAATGAACGGGGTGAAAAACTGATCGAGTTCTGCATTGAAAGAAATCTCAGTATTATGAACACTTACTTCCAACATCATCCAAGACGCTTATACACGTGGAGATCTCCAGGTGACAGGTACAGAAATCAAATCGACTACATCATTCTAGATGGCCGCTGGAGATCCTCAGTATCTAACGTGAAGACCTTTCCCGGCGCAGAGTGCGGCTCCGACCATAACCTGCTCGTAGCTAACTTCTCGCTGCGTTTAAAGGCGCCCAGACGTCAACCACCAAAACCCAATAGTTTGGATCCTTCCGAACAACCGATCTTCCGAGAGGCATTAGAGGCGATCTTGAAGGATTCTCCAACGCACATTCACGCTAACGCTGATACACAGTGGAAACACTTGAAAGGTCACATCAACGATGCGCTGGACCAGATCGCTAAAAAGAAAAAGCCAGAACCGAAGAAAAAATGCTGGATCTCCGATCATTCGTGGGAGCTTATTCAACGCAGGAAAGACTTAAAAACTAGCGGTTTGCAAAATGGTGCAAAGGTTAAATACTCCGAACTTAGCAGAGCTATTCGGGTCAGTTGTCGCCTTGACAAGAACCACTTTGTTGAGAAAAATCGTGCGTGCGAAGGTGATACAAAAGGGTGGGCACGACCCTCAGCAATGAGGAATACGACGCAAGGTGGAGGTAATTATACATACTTATATCATGTATATAAGTCTCAATTAAGGGAGCATTTCAGGGACTTTCACATTTGA
Protein
MESVTTRVNLLGNPTNHRRHPVSGLDGVKLATEASPEVNRQASEGGDTTVGTAASKSEKTSIRSSHRIGAWNVRGLLEAGKLTIVEKEMESHDISILGISETHMRGNGHFTTATGNTMYFSGCEDSSLSGVGIILPPHLNKYVLGYNPVSDRILTMKLNTNPCVLNVIQIYAPTAQAQDELIVDFYGKLEEILNSIPSREIKIILGDWNAKVGSTLDDNHIRSTVGKFGLGIRNERGEKLIEFCIERNLSIMNTYFQHHPRRLYTWRSPGDRYRNQIDYIILDGRWRSSVSNVKTFPGAECGSDHNLLVANFSLRLKAPRRQPPKPNSLDPSEQPIFREALEAILKDSPTHIHANADTQWKHLKGHINDALDQIAKKKKPEPKKKCWISDHSWELIQRRKDLKTSGLQNGAKVKYSELSRAIRVSCRLDKNHFVEKNRACEGDTKGWARPSAMRNTTQGGGNYTYLYHVYKSQLREHFRDFHI
Summary
Uniprot
A0A2H1W4B8
D7F168
A0A2W1BNH4
A0A2W1BHL0
D7F163
D7F172
+ More
D7F161 A0A0J7L4A4 D7F174 D5LB39 D7F162 D7F175 A0A2S2NUN8 D7F167 A0A2G8JV65 A0A3Q1M0C5 A0A3B1JJK0 A0A3B1JKT5 A0A3Q1NNM3 A0A3Q1LMM1 A0A023GD30 A0A3S3P648 J9JN90 A0A2J7QBY6 A0A0B7BR36 A0A0B7BRV7 A0A2J7RSI1 A0A2J7RP40 A0A3Q1MSH0 A0A2J7R2Y0 A0A3Q1NFT8 A0A2J7Q4I0 A0A3Q1M255 A0A2J7PTB8 A0A2J7PFC5 A0A2J7PSI0 A0A1S3DNU9 A0A2J7PSH5 A0A2J7QEJ0 A0A2J7QI56 A0A2J7PQA3 A0A2J7Q6T5 A0A2J7RLI7 A0A2J7RGX2 A0A2J7QGX8 A0A2J7RP47 A0A2J7PHM9 A0A2J7QGJ2 A0A2J7QUN7 A0A2J7QGT4 A0A2J7R4R4 A0A2J7Q3T8 A0A2J7QH51 A0A2J7Q0C6 A0A2J7Q5D5 A0A2J7QKB0 A0A2J7Q2Z3 A0A2J7PMJ3 W4YZ24
D7F161 A0A0J7L4A4 D7F174 D5LB39 D7F162 D7F175 A0A2S2NUN8 D7F167 A0A2G8JV65 A0A3Q1M0C5 A0A3B1JJK0 A0A3B1JKT5 A0A3Q1NNM3 A0A3Q1LMM1 A0A023GD30 A0A3S3P648 J9JN90 A0A2J7QBY6 A0A0B7BR36 A0A0B7BRV7 A0A2J7RSI1 A0A2J7RP40 A0A3Q1MSH0 A0A2J7R2Y0 A0A3Q1NFT8 A0A2J7Q4I0 A0A3Q1M255 A0A2J7PTB8 A0A2J7PFC5 A0A2J7PSI0 A0A1S3DNU9 A0A2J7PSH5 A0A2J7QEJ0 A0A2J7QI56 A0A2J7PQA3 A0A2J7Q6T5 A0A2J7RLI7 A0A2J7RGX2 A0A2J7QGX8 A0A2J7RP47 A0A2J7PHM9 A0A2J7QGJ2 A0A2J7QUN7 A0A2J7QGT4 A0A2J7R4R4 A0A2J7Q3T8 A0A2J7QH51 A0A2J7Q0C6 A0A2J7Q5D5 A0A2J7QKB0 A0A2J7Q2Z3 A0A2J7PMJ3 W4YZ24
EMBL
ODYU01006118
SOQ47682.1
FJ265553
ADI61821.1
KZ150025
PZC74827.1
+ More
KZ150087 PZC73751.1 FJ265548 ADI61816.1 FJ265557 ADI61825.1 FJ265546 ADI61814.1 LBMM01000801 KMQ97461.1 FJ265559 ADI61827.1 GU815090 ADF18553.1 FJ265547 ADI61815.1 FJ265560 ADI61828.1 GGMR01007827 MBY20446.1 FJ265552 ADI61820.1 MRZV01001216 PIK39651.1 GBBM01003639 JAC31779.1 NCKU01004395 RWS06003.1 ABLF02027064 ABLF02027066 NEVH01016294 PNF26085.1 HACG01048879 CEK95744.1 HACG01048873 CEK95738.1 NEVH01000258 PNF43784.1 NEVH01002141 PNF42594.1 NEVH01007824 PNF35184.1 NEVH01018380 PNF23492.1 NEVH01021610 PNF19569.1 NEVH01025668 PNF15041.1 NEVH01021928 PNF19298.1 NEVH01021931 PNF19256.1 NEVH01015309 PNF27009.1 NEVH01013959 PNF28275.1 NEVH01022640 NEVH01008218 NEVH01005885 PNF18513.1 PNF34430.1 PNF38481.1 NEVH01017452 PNF24293.1 NEVH01002684 PNF41704.1 NEVH01003750 PNF40091.1 NEVH01014359 PNF27813.1 PNF42599.1 NEVH01025135 PNF15828.1 NEVH01018372 NEVH01014371 NEVH01011208 NEVH01002697 NEVH01002552 PNF23678.1 PNF27673.1 PNF31697.1 PNF41592.1 PNF42118.1 NEVH01023960 NEVH01010501 PNF17732.1 PNF32290.1 PNF27789.1 NEVH01007402 PNF35815.1 NEVH01019064 PNF23230.1 NEVH01014358 PNF27914.1 NEVH01019971 PNF22037.1 NEVH01017570 PNF23798.1 NEVH01013275 PNF29009.1 NEVH01019075 PNF22955.1 NEVH01023985 PNF17546.1 AAGJ04003947
KZ150087 PZC73751.1 FJ265548 ADI61816.1 FJ265557 ADI61825.1 FJ265546 ADI61814.1 LBMM01000801 KMQ97461.1 FJ265559 ADI61827.1 GU815090 ADF18553.1 FJ265547 ADI61815.1 FJ265560 ADI61828.1 GGMR01007827 MBY20446.1 FJ265552 ADI61820.1 MRZV01001216 PIK39651.1 GBBM01003639 JAC31779.1 NCKU01004395 RWS06003.1 ABLF02027064 ABLF02027066 NEVH01016294 PNF26085.1 HACG01048879 CEK95744.1 HACG01048873 CEK95738.1 NEVH01000258 PNF43784.1 NEVH01002141 PNF42594.1 NEVH01007824 PNF35184.1 NEVH01018380 PNF23492.1 NEVH01021610 PNF19569.1 NEVH01025668 PNF15041.1 NEVH01021928 PNF19298.1 NEVH01021931 PNF19256.1 NEVH01015309 PNF27009.1 NEVH01013959 PNF28275.1 NEVH01022640 NEVH01008218 NEVH01005885 PNF18513.1 PNF34430.1 PNF38481.1 NEVH01017452 PNF24293.1 NEVH01002684 PNF41704.1 NEVH01003750 PNF40091.1 NEVH01014359 PNF27813.1 PNF42599.1 NEVH01025135 PNF15828.1 NEVH01018372 NEVH01014371 NEVH01011208 NEVH01002697 NEVH01002552 PNF23678.1 PNF27673.1 PNF31697.1 PNF41592.1 PNF42118.1 NEVH01023960 NEVH01010501 PNF17732.1 PNF32290.1 PNF27789.1 NEVH01007402 PNF35815.1 NEVH01019064 PNF23230.1 NEVH01014358 PNF27914.1 NEVH01019971 PNF22037.1 NEVH01017570 PNF23798.1 NEVH01013275 PNF29009.1 NEVH01019075 PNF22955.1 NEVH01023985 PNF17546.1 AAGJ04003947
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
A0A2H1W4B8
D7F168
A0A2W1BNH4
A0A2W1BHL0
D7F163
D7F172
+ More
D7F161 A0A0J7L4A4 D7F174 D5LB39 D7F162 D7F175 A0A2S2NUN8 D7F167 A0A2G8JV65 A0A3Q1M0C5 A0A3B1JJK0 A0A3B1JKT5 A0A3Q1NNM3 A0A3Q1LMM1 A0A023GD30 A0A3S3P648 J9JN90 A0A2J7QBY6 A0A0B7BR36 A0A0B7BRV7 A0A2J7RSI1 A0A2J7RP40 A0A3Q1MSH0 A0A2J7R2Y0 A0A3Q1NFT8 A0A2J7Q4I0 A0A3Q1M255 A0A2J7PTB8 A0A2J7PFC5 A0A2J7PSI0 A0A1S3DNU9 A0A2J7PSH5 A0A2J7QEJ0 A0A2J7QI56 A0A2J7PQA3 A0A2J7Q6T5 A0A2J7RLI7 A0A2J7RGX2 A0A2J7QGX8 A0A2J7RP47 A0A2J7PHM9 A0A2J7QGJ2 A0A2J7QUN7 A0A2J7QGT4 A0A2J7R4R4 A0A2J7Q3T8 A0A2J7QH51 A0A2J7Q0C6 A0A2J7Q5D5 A0A2J7QKB0 A0A2J7Q2Z3 A0A2J7PMJ3 W4YZ24
D7F161 A0A0J7L4A4 D7F174 D5LB39 D7F162 D7F175 A0A2S2NUN8 D7F167 A0A2G8JV65 A0A3Q1M0C5 A0A3B1JJK0 A0A3B1JKT5 A0A3Q1NNM3 A0A3Q1LMM1 A0A023GD30 A0A3S3P648 J9JN90 A0A2J7QBY6 A0A0B7BR36 A0A0B7BRV7 A0A2J7RSI1 A0A2J7RP40 A0A3Q1MSH0 A0A2J7R2Y0 A0A3Q1NFT8 A0A2J7Q4I0 A0A3Q1M255 A0A2J7PTB8 A0A2J7PFC5 A0A2J7PSI0 A0A1S3DNU9 A0A2J7PSH5 A0A2J7QEJ0 A0A2J7QI56 A0A2J7PQA3 A0A2J7Q6T5 A0A2J7RLI7 A0A2J7RGX2 A0A2J7QGX8 A0A2J7RP47 A0A2J7PHM9 A0A2J7QGJ2 A0A2J7QUN7 A0A2J7QGT4 A0A2J7R4R4 A0A2J7Q3T8 A0A2J7QH51 A0A2J7Q0C6 A0A2J7Q5D5 A0A2J7QKB0 A0A2J7Q2Z3 A0A2J7PMJ3 W4YZ24
Ontologies
PANTHER
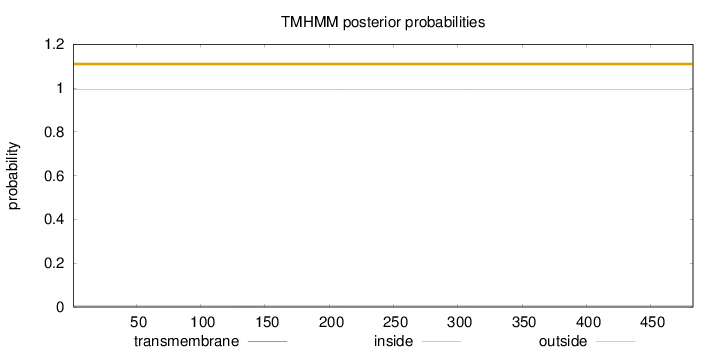
Topology
Length:
483
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0072
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00580
outside
1 - 483
Population Genetic Test Statistics
Pi
470.376017
Theta
228.468884
Tajima's D
4.015384
CLR
0
CSRT
0.998800059997
Interpretation
Possibly Balancing Selection
