Gene
KWMTBOMO13329 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004681
Annotation
alpha-tubulin_[Bombyx_mori]
Full name
Tubulin alpha chain
Alternative Name
Alpha-tubulin
Alpha T6
Alpha T6
Location in the cell
Cytoplasmic Reliability : 2.496
Sequence
CDS
ATGAGGGAGTGCATATCCGTACACGTGGGCCAAGCCGGGGTCCAGATGGGCGTCGCGTGCTGGCAGCTATACTGCTTGGAGCACGGGATCCGACCTGACGGAACCCTGCCCGCTTGTGAAACTGATCCTAATACAGCGGATTCCTGCTTCAATACGTTTTTTTCTGAGGCTGATCGAGGGAAGATGGTGCCGAGGGTCGTGATGGTTGATTTAGAGGCTACTGTTATAGACGAAGTTCGTTCCGGTGAATACCGCCAACTGTATCACCCTGAACAGTTGATAACTGGTAAAGAAGACGCGGCGAATAACTACGCGAGAGGGCACTATTCGACCGGACGCGAGGTTCTCAGTCCGGTTATGGAGCGAATTAGGAAACTTGCCGATCAATGTACCGGACTACAGGGTTTCTTCGTGTTTCATTCGTTTGGAGGTGGCACAGGATCTGGATTCACATCGCTACTGATGGAGAAGCTATCTGACGAGTTTGGAAAGAAGAGTAAACTGGAATTTGCCATATATCCTGCGCCACAAGTGTCAACTGCGGTAGTGGAGCCATACAACGCGGTGCTAACTACGCACGCGACCATCAGTCATTCGGATTGCGCCTTCATGGTGGACAATGAAGCTATCTACGACATTTGCAGGCGAAGGCTGTCTATAGAGAGACCTTCTTATGCAAATTTGAATAGGCTTATCTCACAAGTAGTTTCGTCTATAACGGCTTCACTACGATTCGATGGCGCCCTAAATGTGGATTTGACGGAATTCCAAACGAATCTCGTTCCCTACCCAAGGATACATTTTCCACTAGCCGCGTACGCACCCGTTGTTTCTGCTGATAAGGCGTACCACGAAGGTATGTCTGTGTCGGAGATCACTGCGGAACTGTTCGAGCCCCAGAACCAGATGGTGAAGTGCGATCCTCGAGACGGAAAGTACATGGCGTGCTGCCTGCTGTACAGGGGAGACGTGGTCCCGAAGGACGTGAACGCGGCTATAGCGGCCATGAAGGGCAGGGCCGGAATACGGTTTGTGGATTGGTGTCCTACCGGTTTTAAAGTTGGTATAAACTATCAGCCGCCGTCTGTGGTGACGGGCGGGGATCTGGCCCAAGTGAAGCGGGCCGCCTCTATGCTCAGTAACACCACCGCTATAGCGGAGGCCTGGGGCAAGCTTGACCACAAGTTCGACCTCATGTATTCCAAGCGAGCTTTTGTGCATTGGTATGTAGGCGAAGGGATGGAGGAAGGCGAATTCTCTGAAGCGAGAGAAGATTTGGCAGCTCTGGAACGAGATTACGATGAAGTGGCTATTGAGACGTCAGACATGCAGCCCGGCGCCGATGATGAGTTATGA
Protein
MRECISVHVGQAGVQMGVACWQLYCLEHGIRPDGTLPACETDPNTADSCFNTFFSEADRGKMVPRVVMVDLEATVIDEVRSGEYRQLYHPEQLITGKEDAANNYARGHYSTGREVLSPVMERIRKLADQCTGLQGFFVFHSFGGGTGSGFTSLLMEKLSDEFGKKSKLEFAIYPAPQVSTAVVEPYNAVLTTHATISHSDCAFMVDNEAIYDICRRRLSIERPSYANLNRLISQVVSSITASLRFDGALNVDLTEFQTNLVPYPRIHFPLAAYAPVVSADKAYHEGMSVSEITAELFEPQNQMVKCDPRDGKYMACCLLYRGDVVPKDVNAAIAAMKGRAGIRFVDWCPTGFKVGINYQPPSVVTGGDLAQVKRAASMLSNTTAIAEAWGKLDHKFDLMYSKRAFVHWYVGEGMEEGEFSEAREDLAALERDYDEVAIETSDMQPGADDEL
Summary
Description
Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
Subunit
Dimer of alpha and beta chains. A typical microtubule is a hollow water-filled tube with an outer diameter of 25 nm and an inner diameter of 15 nM. Alpha-beta heterodimers associate head-to-tail to form protofilaments running lengthwise along the microtubule wall with the beta-tubulin subunit facing the microtubule plus end conferring a structural polarity. Microtubules usually have 13 protofilaments but different protofilament numbers can be found in some organisms and specialized cells.
Similarity
Belongs to the tubulin family.
Keywords
Acetylation
Complete proteome
Cytoplasm
Cytoskeleton
GTP-binding
Isopeptide bond
Microtubule
Nucleotide-binding
Reference proteome
Allergen
Feature
chain Tubulin alpha chain
Uniprot
Q8T8B4
A0A2A4JX96
A0A2H1WZY3
A0A3S2NFZ3
H9J591
A0A212EQV5
+ More
A0A194REY2 A0A088AEZ4 A0A2A3ECG0 A0A026X3W6 A0A0L7QL05 A0A3L8DJ67 A0A151I9S0 A0A154PGT8 E2BYM3 A0A0J7KVC1 A0A1Y1MUS0 A0A0M8ZUN7 A0A195FGK2 A0A2P8YMG0 E2A2Y4 W6UDI6 A0A195E3R3 A0A1Y1N000 S7Q4Z3 B7ZTG2 Q28IX8 Q9YHW2 A0A2I4D018 A0A3Q3L447 A0A3P8QCW6 V4A2S7 A0A0K8RDS4 V4CNQ5 R7T6Z8 B7QP06 Q8WQ47 A0A3Q3E9D2 A0A3B3ICA2 A0A3P9IA60 A0A3P8QDU8 Q6DD58 W4ZJ54 A0A3B3YZV5 A0A2I4D484 A0A096M087 A0A3Q3F4W5 A0A3B3VZH0 A0A2R5LLI8 A0A1A9YUF9 A0A1A9XMZ8 A0A1B0C2X1 A0A1Z5KXS7 A0A1B0A8T1 A0A069DZ13 A0A1A9W6N7 A0A0R3WDD2 A0A3Q1JWL4 A0A3P8QDI8 H3AGU2 A0A1Y3BTA3 A0A131XVH2 A0A068J741 A0A3S4R6E6 A0A0R3TZZ1 W5MXH5 A0A3B1JFU0 A0A3P9H4L4 H3AGU3 Q52PV9 A0A226EVU5 V4CGI5 A0A131ZV00 A0A1L8HHQ5 G3PCL3 A0A3N0Z1J9 F6U388 A0A068XPQ2 A0A1I8PKP1 A0A0L0CGN0 P36220 A0A1L8H9K3 A0A3Q2PC85 A0A3B3TDV4 G3PCE1 A0A3B3DK83 A0A3B4TNK3 A0A0B8RPP3 A0A3P9DDJ3 J3SDL3 A0A2U9C1Z1 Q8AWD6 H2MPB8 A0A250YI73 L5LYK7 A0A3Q7SMU8 A0A224XCE3 A0A0P4VZJ1 A0A1A9UJX4 R4G8S7
A0A194REY2 A0A088AEZ4 A0A2A3ECG0 A0A026X3W6 A0A0L7QL05 A0A3L8DJ67 A0A151I9S0 A0A154PGT8 E2BYM3 A0A0J7KVC1 A0A1Y1MUS0 A0A0M8ZUN7 A0A195FGK2 A0A2P8YMG0 E2A2Y4 W6UDI6 A0A195E3R3 A0A1Y1N000 S7Q4Z3 B7ZTG2 Q28IX8 Q9YHW2 A0A2I4D018 A0A3Q3L447 A0A3P8QCW6 V4A2S7 A0A0K8RDS4 V4CNQ5 R7T6Z8 B7QP06 Q8WQ47 A0A3Q3E9D2 A0A3B3ICA2 A0A3P9IA60 A0A3P8QDU8 Q6DD58 W4ZJ54 A0A3B3YZV5 A0A2I4D484 A0A096M087 A0A3Q3F4W5 A0A3B3VZH0 A0A2R5LLI8 A0A1A9YUF9 A0A1A9XMZ8 A0A1B0C2X1 A0A1Z5KXS7 A0A1B0A8T1 A0A069DZ13 A0A1A9W6N7 A0A0R3WDD2 A0A3Q1JWL4 A0A3P8QDI8 H3AGU2 A0A1Y3BTA3 A0A131XVH2 A0A068J741 A0A3S4R6E6 A0A0R3TZZ1 W5MXH5 A0A3B1JFU0 A0A3P9H4L4 H3AGU3 Q52PV9 A0A226EVU5 V4CGI5 A0A131ZV00 A0A1L8HHQ5 G3PCL3 A0A3N0Z1J9 F6U388 A0A068XPQ2 A0A1I8PKP1 A0A0L0CGN0 P36220 A0A1L8H9K3 A0A3Q2PC85 A0A3B3TDV4 G3PCE1 A0A3B3DK83 A0A3B4TNK3 A0A0B8RPP3 A0A3P9DDJ3 J3SDL3 A0A2U9C1Z1 Q8AWD6 H2MPB8 A0A250YI73 L5LYK7 A0A3Q7SMU8 A0A224XCE3 A0A0P4VZJ1 A0A1A9UJX4 R4G8S7
Pubmed
EMBL
AB072306
BAB86851.1
NWSH01000459
PCG76254.1
ODYU01012310
SOQ58562.1
+ More
RSAL01000125 RVE46588.1 BABH01039687 AGBW02013197 OWR43883.1 KQ460323 KPJ15855.1 KZ288287 PBC29425.1 KK107019 EZA62783.1 KQ414940 KOC59211.1 QOIP01000007 RLU20474.1 KQ978268 KYM95822.1 KQ434902 KZC11096.1 GL451499 EFN79177.1 LBMM01002880 KMQ94251.1 GEZM01021611 JAV88908.1 KQ435879 KOX69879.1 KQ981606 KYN39523.1 PYGN01000491 PSN45442.1 GL436239 EFN72192.1 APAU02000118 EUB56387.1 KQ979685 KYN19815.1 GEZM01021612 JAV88907.1 KE164532 EPQ18458.1 BC170862 BC171015 AAI70862.1 CR760170 AF082027 AAC97928.1 KB201263 ESO98168.1 GADI01005204 JAA68604.1 KB199905 ESP04035.1 AMQN01014879 KB311390 ELT89394.1 ABJB010009611 ABJB010251127 DS980981 EEC20578.1 AJ428050 BC077769 CM004482 AAH77769.1 OCT63377.1 AAGJ04114327 AYCK01016441 GGLE01006061 MBY10187.1 CCAG010023980 CCAG010023981 JXJN01024715 JXJN01024716 GFJQ02007027 JAV99942.1 GBGD01001440 JAC87449.1 UYRS01018887 VDK40999.1 AFYH01201289 AFYH01201290 AFYH01201291 MUJZ01000105 OTF84219.1 GEFM01005474 JAP70322.1 KJ690265 AIE13824.1 NCKU01001410 NCKU01001402 RWS12163.1 RWS12179.1 UZAE01015393 VDO15853.1 AHAT01034396 AHAT01034397 AY986760 AAX84656.1 LNIX01000002 OXA60971.1 KB200522 ESP01200.1 JXLN01002314 KPM02593.1 CM004468 OCT95640.1 RJVU01017312 ROL52161.1 JRES01000426 KNC31385.1 X71980 CM004469 OCT92773.1 GBSH01002947 JAG66080.1 JU176342 AFJ51865.1 CP026253 AWP10073.1 BC042319 BC165818 AAH42319.1 AAI65818.1 GFFW01001461 JAV43327.1 KB106857 ELK30543.1 GFTR01006330 JAW10096.1 GDKW01000590 JAI56005.1 GAHY01000452 JAA77058.1
RSAL01000125 RVE46588.1 BABH01039687 AGBW02013197 OWR43883.1 KQ460323 KPJ15855.1 KZ288287 PBC29425.1 KK107019 EZA62783.1 KQ414940 KOC59211.1 QOIP01000007 RLU20474.1 KQ978268 KYM95822.1 KQ434902 KZC11096.1 GL451499 EFN79177.1 LBMM01002880 KMQ94251.1 GEZM01021611 JAV88908.1 KQ435879 KOX69879.1 KQ981606 KYN39523.1 PYGN01000491 PSN45442.1 GL436239 EFN72192.1 APAU02000118 EUB56387.1 KQ979685 KYN19815.1 GEZM01021612 JAV88907.1 KE164532 EPQ18458.1 BC170862 BC171015 AAI70862.1 CR760170 AF082027 AAC97928.1 KB201263 ESO98168.1 GADI01005204 JAA68604.1 KB199905 ESP04035.1 AMQN01014879 KB311390 ELT89394.1 ABJB010009611 ABJB010251127 DS980981 EEC20578.1 AJ428050 BC077769 CM004482 AAH77769.1 OCT63377.1 AAGJ04114327 AYCK01016441 GGLE01006061 MBY10187.1 CCAG010023980 CCAG010023981 JXJN01024715 JXJN01024716 GFJQ02007027 JAV99942.1 GBGD01001440 JAC87449.1 UYRS01018887 VDK40999.1 AFYH01201289 AFYH01201290 AFYH01201291 MUJZ01000105 OTF84219.1 GEFM01005474 JAP70322.1 KJ690265 AIE13824.1 NCKU01001410 NCKU01001402 RWS12163.1 RWS12179.1 UZAE01015393 VDO15853.1 AHAT01034396 AHAT01034397 AY986760 AAX84656.1 LNIX01000002 OXA60971.1 KB200522 ESP01200.1 JXLN01002314 KPM02593.1 CM004468 OCT95640.1 RJVU01017312 ROL52161.1 JRES01000426 KNC31385.1 X71980 CM004469 OCT92773.1 GBSH01002947 JAG66080.1 JU176342 AFJ51865.1 CP026253 AWP10073.1 BC042319 BC165818 AAH42319.1 AAI65818.1 GFFW01001461 JAV43327.1 KB106857 ELK30543.1 GFTR01006330 JAW10096.1 GDKW01000590 JAI56005.1 GAHY01000452 JAA77058.1
Proteomes
UP000218220
UP000283053
UP000005204
UP000007151
UP000053240
UP000005203
+ More
UP000242457 UP000053097 UP000053825 UP000279307 UP000078542 UP000076502 UP000008237 UP000036403 UP000053105 UP000078541 UP000245037 UP000000311 UP000019149 UP000078492 UP000008143 UP000192220 UP000261660 UP000265100 UP000030746 UP000014760 UP000001555 UP000001038 UP000265200 UP000186698 UP000007110 UP000261480 UP000028760 UP000264800 UP000261500 UP000092444 UP000092443 UP000092460 UP000092445 UP000091820 UP000046400 UP000282613 UP000265040 UP000008672 UP000285301 UP000046398 UP000278807 UP000018468 UP000018467 UP000198287 UP000007635 UP000002280 UP000095300 UP000037069 UP000265000 UP000261540 UP000261560 UP000261420 UP000265160 UP000246464 UP000286640 UP000078200
UP000242457 UP000053097 UP000053825 UP000279307 UP000078542 UP000076502 UP000008237 UP000036403 UP000053105 UP000078541 UP000245037 UP000000311 UP000019149 UP000078492 UP000008143 UP000192220 UP000261660 UP000265100 UP000030746 UP000014760 UP000001555 UP000001038 UP000265200 UP000186698 UP000007110 UP000261480 UP000028760 UP000264800 UP000261500 UP000092444 UP000092443 UP000092460 UP000092445 UP000091820 UP000046400 UP000282613 UP000265040 UP000008672 UP000285301 UP000046398 UP000278807 UP000018468 UP000018467 UP000198287 UP000007635 UP000002280 UP000095300 UP000037069 UP000265000 UP000261540 UP000261560 UP000261420 UP000265160 UP000246464 UP000286640 UP000078200
Pfam
Interpro
IPR036525
Tubulin/FtsZ_GTPase_sf
+ More
IPR023123 Tubulin_C
IPR000217 Tubulin
IPR018316 Tubulin/FtsZ_2-layer-sand-dom
IPR003008 Tubulin_FtsZ_GTPase
IPR008280 Tub_FtsZ_C
IPR037103 Tubulin/FtsZ_C_sf
IPR002452 Alpha_tubulin
IPR017975 Tubulin_CS
IPR017901 C-CAP_CF_C-like
IPR006786 Pinin_SDK_MemA
IPR012945 Tubulin-bd_cofactor_C_dom
IPR031925 TBCC_N
IPR016098 CAP/MinC_C
IPR006599 CARP_motif
IPR038397 TBCC_N_sf
IPR002699 V_ATPase_D
IPR029004 L28e/Mak16
IPR023123 Tubulin_C
IPR000217 Tubulin
IPR018316 Tubulin/FtsZ_2-layer-sand-dom
IPR003008 Tubulin_FtsZ_GTPase
IPR008280 Tub_FtsZ_C
IPR037103 Tubulin/FtsZ_C_sf
IPR002452 Alpha_tubulin
IPR017975 Tubulin_CS
IPR017901 C-CAP_CF_C-like
IPR006786 Pinin_SDK_MemA
IPR012945 Tubulin-bd_cofactor_C_dom
IPR031925 TBCC_N
IPR016098 CAP/MinC_C
IPR006599 CARP_motif
IPR038397 TBCC_N_sf
IPR002699 V_ATPase_D
IPR029004 L28e/Mak16
ProteinModelPortal
Q8T8B4
A0A2A4JX96
A0A2H1WZY3
A0A3S2NFZ3
H9J591
A0A212EQV5
+ More
A0A194REY2 A0A088AEZ4 A0A2A3ECG0 A0A026X3W6 A0A0L7QL05 A0A3L8DJ67 A0A151I9S0 A0A154PGT8 E2BYM3 A0A0J7KVC1 A0A1Y1MUS0 A0A0M8ZUN7 A0A195FGK2 A0A2P8YMG0 E2A2Y4 W6UDI6 A0A195E3R3 A0A1Y1N000 S7Q4Z3 B7ZTG2 Q28IX8 Q9YHW2 A0A2I4D018 A0A3Q3L447 A0A3P8QCW6 V4A2S7 A0A0K8RDS4 V4CNQ5 R7T6Z8 B7QP06 Q8WQ47 A0A3Q3E9D2 A0A3B3ICA2 A0A3P9IA60 A0A3P8QDU8 Q6DD58 W4ZJ54 A0A3B3YZV5 A0A2I4D484 A0A096M087 A0A3Q3F4W5 A0A3B3VZH0 A0A2R5LLI8 A0A1A9YUF9 A0A1A9XMZ8 A0A1B0C2X1 A0A1Z5KXS7 A0A1B0A8T1 A0A069DZ13 A0A1A9W6N7 A0A0R3WDD2 A0A3Q1JWL4 A0A3P8QDI8 H3AGU2 A0A1Y3BTA3 A0A131XVH2 A0A068J741 A0A3S4R6E6 A0A0R3TZZ1 W5MXH5 A0A3B1JFU0 A0A3P9H4L4 H3AGU3 Q52PV9 A0A226EVU5 V4CGI5 A0A131ZV00 A0A1L8HHQ5 G3PCL3 A0A3N0Z1J9 F6U388 A0A068XPQ2 A0A1I8PKP1 A0A0L0CGN0 P36220 A0A1L8H9K3 A0A3Q2PC85 A0A3B3TDV4 G3PCE1 A0A3B3DK83 A0A3B4TNK3 A0A0B8RPP3 A0A3P9DDJ3 J3SDL3 A0A2U9C1Z1 Q8AWD6 H2MPB8 A0A250YI73 L5LYK7 A0A3Q7SMU8 A0A224XCE3 A0A0P4VZJ1 A0A1A9UJX4 R4G8S7
A0A194REY2 A0A088AEZ4 A0A2A3ECG0 A0A026X3W6 A0A0L7QL05 A0A3L8DJ67 A0A151I9S0 A0A154PGT8 E2BYM3 A0A0J7KVC1 A0A1Y1MUS0 A0A0M8ZUN7 A0A195FGK2 A0A2P8YMG0 E2A2Y4 W6UDI6 A0A195E3R3 A0A1Y1N000 S7Q4Z3 B7ZTG2 Q28IX8 Q9YHW2 A0A2I4D018 A0A3Q3L447 A0A3P8QCW6 V4A2S7 A0A0K8RDS4 V4CNQ5 R7T6Z8 B7QP06 Q8WQ47 A0A3Q3E9D2 A0A3B3ICA2 A0A3P9IA60 A0A3P8QDU8 Q6DD58 W4ZJ54 A0A3B3YZV5 A0A2I4D484 A0A096M087 A0A3Q3F4W5 A0A3B3VZH0 A0A2R5LLI8 A0A1A9YUF9 A0A1A9XMZ8 A0A1B0C2X1 A0A1Z5KXS7 A0A1B0A8T1 A0A069DZ13 A0A1A9W6N7 A0A0R3WDD2 A0A3Q1JWL4 A0A3P8QDI8 H3AGU2 A0A1Y3BTA3 A0A131XVH2 A0A068J741 A0A3S4R6E6 A0A0R3TZZ1 W5MXH5 A0A3B1JFU0 A0A3P9H4L4 H3AGU3 Q52PV9 A0A226EVU5 V4CGI5 A0A131ZV00 A0A1L8HHQ5 G3PCL3 A0A3N0Z1J9 F6U388 A0A068XPQ2 A0A1I8PKP1 A0A0L0CGN0 P36220 A0A1L8H9K3 A0A3Q2PC85 A0A3B3TDV4 G3PCE1 A0A3B3DK83 A0A3B4TNK3 A0A0B8RPP3 A0A3P9DDJ3 J3SDL3 A0A2U9C1Z1 Q8AWD6 H2MPB8 A0A250YI73 L5LYK7 A0A3Q7SMU8 A0A224XCE3 A0A0P4VZJ1 A0A1A9UJX4 R4G8S7
PDB
6QTN
E-value=0,
Score=1878
Ontologies
GO
PANTHER
Topology
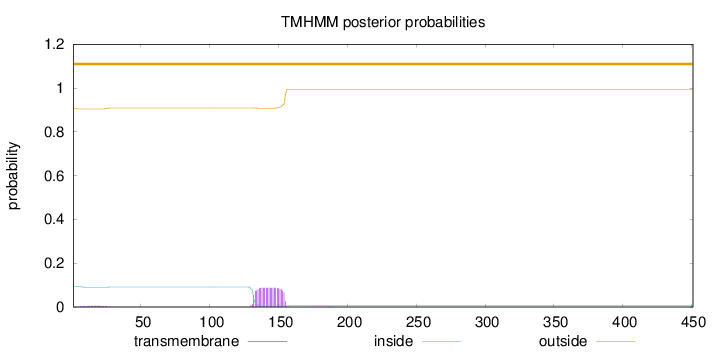
Subcellular location
Length:
451
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.04265
Exp number, first 60 AAs:
0.09388
Total prob of N-in:
0.09499
outside
1 - 451
Population Genetic Test Statistics
Pi
20.261909
Theta
29.906205
Tajima's D
-0.660757
CLR
0.084663
CSRT
0.203089845507725
Interpretation
Uncertain
