Gene
KWMTBOMO13327
Pre Gene Modal
BGIBMGA004679
Annotation
putative_transposase_yabusame-1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.776
Sequence
CDS
ATGGATATTGAAAGACAAGAAGAGCGAATCCGTGCCATGTTAGAGGAGGAGCTAAGTGATTATTCTGATGAATCCAGTTCAGAAGATGAAACCGATCACTGTAGTGAGCACGAAGTGAATTATGACACAGAAGAAGAGCGGATAGATTCAGTTGATGTTCCATCCAACTCCAGGCAGGAAGAAGCAAATGCAATAATAGCAAACGAATCAGATAGTGACCCCGATGACGATTTACCATTGTCTCTCGTCAGGCAGAGAGCTTCAGCTTCTCGACAAGTGAGTGGCCCTTTTTATACGTCAAAAGACGGTACCAAGTGGTATAAAAATTGTCAACGTCCAAACGTAAGGCTTCGTTCCGAAAACATCGTAACAGAGCAAGCTCAAGTAAAAAATATTGCAAGGGATGCTTCTACAGAGTATGAATGCTGGAATATTTTTGTCACTTCAGATATGCTACAAGAAATTTTGACCCACACTAATTCGTCAATTCGCCATAGACAAACCAAAACTGCTGCTGAAAATTCGAGTGCTGAAACTTCTTTCTACATGCAAGAAACCACATTATGTGAATTGAAAGCATTAATTGCATTATTGTACTTAGCTGGCCTTATCAAATCTAATCGACAGAGTTTGAAAGATCTTTGGCGAACCGATGGAACTGGCGTTGATATATTTCGCACCACAATGTCACTCCAGAGATTTCAGTTTCTGCAAAATAATATTCGTTTTGATGACAAATCTACGAGAGATGAGCGTAAACAAACTGATAATATGGCTGCCTTCAGAAGCATTTTTGACCAGTTTGTTCAATGCTGTCAGAATGCGTACTCCCCAAGTGAGTTTCTGACTATCGATGAGATGCTACTATCATTTAGAGGTCGATGCCTGTTTCGTGTCTACATTCCAAATAAACCAGCCAAGTACGGTATAAAAATTTTGGCTCTAGTCGATGCAAAAAACTTTTATGTCGTGAATTTGGAAGTGTACGCAGGAAAACAACCGTCGGGACCATATGCAGTTAGTAATAGACCCTTTGAAGTAGTGGAGAGACTCATTCAACCAGTTGCACGATCACATCGCAACGTGACATTCGATAATTGGTTCACCGGATATGAGCTAATGCTGCATCTACTAAATGAGTATCGACTCACGTCGGTTGGAACTGTACGAAAAAACAAGAGACAGATTCCCGAATCTTTCATTAGAACTGACAGACAACCTAACAGTTCAGTCTTTGGGTTTCAAAAAGATATTACACTCGTTTCCTATGCGCCAAAGAAGAACAAGGTGGTTGTTGTTATGTCCACCATGCATCATGATAACTCAATAGATGAATCTACAGGAGAGAAACAAAAACCTGAAATGATAACATTCTATAATTCGACCAAAGCTGGCGTTGATGTGGTTGATGAGTTGTGTGCCAATTACAACGTTTCTCGCAACAGTAAACGATGGCCTATGACTTTATTCTATGGTGTCTTGAACATGGCAGCTATAAATGCTTGCATTATTTATCGAGCAAATAAAAACGTTACGATCAAGAGAACAGAATTCATTCGTAGTTTGGGCTTATCAATGATTTACGAACATCTGCATTCAAGAAATAAAAAGAAAAATATTCCCACCTACCTCCGCCAGCGAATCGAGAAACAGCTAGGTGAACCATCACCAAGACATGTGAATGTACCTGGGCGATATGTCAGATGCCAGGATTGTCCATACAAGAAGGATAGAAAAACCAAGCATTCCTGCAATGCTTGTGCCAAGCCCATATGTATGGAGCACGCAAAATTTCTTTGCGAAAATTGTGCTGAACTCGACAGTAGTCTTTGA
Protein
MDIERQEERIRAMLEEELSDYSDESSSEDETDHCSEHEVNYDTEEERIDSVDVPSNSRQEEANAIIANESDSDPDDDLPLSLVRQRASASRQVSGPFYTSKDGTKWYKNCQRPNVRLRSENIVTEQAQVKNIARDASTEYECWNIFVTSDMLQEILTHTNSSIRHRQTKTAAENSSAETSFYMQETTLCELKALIALLYLAGLIKSNRQSLKDLWRTDGTGVDIFRTTMSLQRFQFLQNNIRFDDKSTRDERKQTDNMAAFRSIFDQFVQCCQNAYSPSEFLTIDEMLLSFRGRCLFRVYIPNKPAKYGIKILALVDAKNFYVVNLEVYAGKQPSGPYAVSNRPFEVVERLIQPVARSHRNVTFDNWFTGYELMLHLLNEYRLTSVGTVRKNKRQIPESFIRTDRQPNSSVFGFQKDITLVSYAPKKNKVVVVMSTMHHDNSIDESTGEKQKPEMITFYNSTKAGVDVVDELCANYNVSRNSKRWPMTLFYGVLNMAAINACIIYRANKNVTIKRTEFIRSLGLSMIYEHLHSRNKKKNIPTYLRQRIEKQLGEPSPRHVNVPGRYVRCQDCPYKKDRKTKHSCNACAKPICMEHAKFLCENCAELDSSL
Summary
Uniprot
Q75QC0
Q75R41
H9J589
A0A286Y2W0
A0A2A4J0M2
A0A2H1WQI5
+ More
A0A2A4J2P3 J9JSM6 A0A1B6BXA3 A0A1B6E438 A0A1B6JQP1 A0A087T137 A0A1Y1MSE2 X1WKK7 A0A1B6DER6 A0A0V1CRV4 A0A1Y1K4Z8 X1X571 A0A0V1CUA7 X1WSB4 A0A1B6E2X5 J9M3V0 X1WQE2 A0A0V1NJ43 A0A1B6MTB6 A0A087UAY4 A0A1B6LLL4 J9JUF3 Q207U0 A0A1B6KGD1 A0A1B6M803 E7BXG0 X1WSQ3 A0A1B6MIG5 A0A1B6DJR9 A0A0V1N525 A0A1B6MBV4 A0A023F1Y2 A0A0V1LC53 A0A2P8ZLW3 A0A087UHP8 A0A2P8XIJ0 A0A1Y1LGT7 A0A1B6LVC4 A0A0V0W8G5 A0A087TGK5 A0A1W4WUL1 A0A023F5Y4 A0A0K2TIZ0 A0A0V0SH12 A0A1B6CEU0 A0A1B6MC64 J9LFP4 A0A2P8Z261 A0A2P8YU38 A0A2H1V9L6 A0A023F5Y0 B0XHV3 A0A2S2R2M4 A0A2A4K028 A0A087U919 A0A087TH18 X1X9R9 J9JNY3 A0A1B6LUY7 A0A2R7WGP5 A0A146KVM3 A0A2P8XRS2 A0A3L8DT18 A0A1B6C4A1 A0A1B6CPZ3 D2XZW7 B1P5D5
A0A2A4J2P3 J9JSM6 A0A1B6BXA3 A0A1B6E438 A0A1B6JQP1 A0A087T137 A0A1Y1MSE2 X1WKK7 A0A1B6DER6 A0A0V1CRV4 A0A1Y1K4Z8 X1X571 A0A0V1CUA7 X1WSB4 A0A1B6E2X5 J9M3V0 X1WQE2 A0A0V1NJ43 A0A1B6MTB6 A0A087UAY4 A0A1B6LLL4 J9JUF3 Q207U0 A0A1B6KGD1 A0A1B6M803 E7BXG0 X1WSQ3 A0A1B6MIG5 A0A1B6DJR9 A0A0V1N525 A0A1B6MBV4 A0A023F1Y2 A0A0V1LC53 A0A2P8ZLW3 A0A087UHP8 A0A2P8XIJ0 A0A1Y1LGT7 A0A1B6LVC4 A0A0V0W8G5 A0A087TGK5 A0A1W4WUL1 A0A023F5Y4 A0A0K2TIZ0 A0A0V0SH12 A0A1B6CEU0 A0A1B6MC64 J9LFP4 A0A2P8Z261 A0A2P8YU38 A0A2H1V9L6 A0A023F5Y0 B0XHV3 A0A2S2R2M4 A0A2A4K028 A0A087U919 A0A087TH18 X1X9R9 J9JNY3 A0A1B6LUY7 A0A2R7WGP5 A0A146KVM3 A0A2P8XRS2 A0A3L8DT18 A0A1B6C4A1 A0A1B6CPZ3 D2XZW7 B1P5D5
EMBL
AB162707
BAD11135.1
AB159601
BAD07480.1
BABH01039687
AAKN02047000
+ More
NWSH01004271 PCG65286.1 ODYU01010244 SOQ55222.1 NWSH01003816 PCG65794.1 ABLF02012085 ABLF02012086 GEDC01031381 JAS05917.1 GEDC01007104 GEDC01004639 JAS30194.1 JAS32659.1 GECU01006493 JAT01214.1 KK112917 KFM58826.1 GEZM01022990 GEZM01022989 JAV88624.1 ABLF02025070 GEDC01013105 JAS24193.1 JYDI01000114 KRY51933.1 GEZM01092454 JAV56552.1 ABLF02010724 JYDI01000099 KRY52767.1 ABLF02014529 GEDC01005015 JAS32283.1 ABLF02010723 ABLF02061909 JYDM01000192 KRZ83916.1 GEBQ01000795 JAT39182.1 KK119053 KFM74523.1 GEBQ01015347 JAT24630.1 ABLF02002291 ABLF02056818 DQ407726 ABD76335.1 GEBQ01029484 JAT10493.1 GEBQ01008003 JAT31974.1 GU329918 ADU04477.1 ABLF02003601 ABLF02054890 GEBQ01004274 JAT35703.1 GEDC01011391 JAS25907.1 JYDO01000008 KRZ79105.1 GEBQ01006564 JAT33413.1 GBBI01003661 JAC15051.1 JYDW01000090 KRZ56622.1 PYGN01000020 PSN57460.1 KK119845 KFM76887.1 PYGN01002011 PYGN01000918 PSN31819.1 PSN39358.1 GEZM01056111 JAV72843.1 GEBQ01012372 JAT27605.1 JYDK01000225 KRX71861.1 KK115134 KFM64244.1 GBBI01002117 JAC16595.1 HACA01008647 CDW26008.1 JYDL01000009 KRX26044.1 GEDC01025350 JAS11948.1 GEBQ01006503 JAT33474.1 ABLF02019776 ABLF02019781 PYGN01000233 PSN50578.1 PYGN01000358 PSN47763.1 ODYU01001398 SOQ37519.1 GBBI01002118 JAC16594.1 DS233215 EDS28762.1 GGMS01014827 MBY84030.1 NWSH01000300 PCG77621.1 KK118795 KFM73858.1 KK115202 KFM64407.1 ABLF02009521 ABLF02003596 GEBQ01012494 JAT27483.1 KK854797 PTY18824.1 GDHC01018085 JAQ00544.1 PYGN01001458 PSN34708.1 QOIP01000004 RLU23313.1 GEDC01029238 JAS08060.1 GEDC01021722 JAS15576.1 GU270322 ADB45159.1 EU287451 ABZ85926.1
NWSH01004271 PCG65286.1 ODYU01010244 SOQ55222.1 NWSH01003816 PCG65794.1 ABLF02012085 ABLF02012086 GEDC01031381 JAS05917.1 GEDC01007104 GEDC01004639 JAS30194.1 JAS32659.1 GECU01006493 JAT01214.1 KK112917 KFM58826.1 GEZM01022990 GEZM01022989 JAV88624.1 ABLF02025070 GEDC01013105 JAS24193.1 JYDI01000114 KRY51933.1 GEZM01092454 JAV56552.1 ABLF02010724 JYDI01000099 KRY52767.1 ABLF02014529 GEDC01005015 JAS32283.1 ABLF02010723 ABLF02061909 JYDM01000192 KRZ83916.1 GEBQ01000795 JAT39182.1 KK119053 KFM74523.1 GEBQ01015347 JAT24630.1 ABLF02002291 ABLF02056818 DQ407726 ABD76335.1 GEBQ01029484 JAT10493.1 GEBQ01008003 JAT31974.1 GU329918 ADU04477.1 ABLF02003601 ABLF02054890 GEBQ01004274 JAT35703.1 GEDC01011391 JAS25907.1 JYDO01000008 KRZ79105.1 GEBQ01006564 JAT33413.1 GBBI01003661 JAC15051.1 JYDW01000090 KRZ56622.1 PYGN01000020 PSN57460.1 KK119845 KFM76887.1 PYGN01002011 PYGN01000918 PSN31819.1 PSN39358.1 GEZM01056111 JAV72843.1 GEBQ01012372 JAT27605.1 JYDK01000225 KRX71861.1 KK115134 KFM64244.1 GBBI01002117 JAC16595.1 HACA01008647 CDW26008.1 JYDL01000009 KRX26044.1 GEDC01025350 JAS11948.1 GEBQ01006503 JAT33474.1 ABLF02019776 ABLF02019781 PYGN01000233 PSN50578.1 PYGN01000358 PSN47763.1 ODYU01001398 SOQ37519.1 GBBI01002118 JAC16594.1 DS233215 EDS28762.1 GGMS01014827 MBY84030.1 NWSH01000300 PCG77621.1 KK118795 KFM73858.1 KK115202 KFM64407.1 ABLF02009521 ABLF02003596 GEBQ01012494 JAT27483.1 KK854797 PTY18824.1 GDHC01018085 JAQ00544.1 PYGN01001458 PSN34708.1 QOIP01000004 RLU23313.1 GEDC01029238 JAS08060.1 GEDC01021722 JAS15576.1 GU270322 ADB45159.1 EU287451 ABZ85926.1
Proteomes
Interpro
SUPFAM
SSF118310
SSF118310
Gene 3D
ProteinModelPortal
Q75QC0
Q75R41
H9J589
A0A286Y2W0
A0A2A4J0M2
A0A2H1WQI5
+ More
A0A2A4J2P3 J9JSM6 A0A1B6BXA3 A0A1B6E438 A0A1B6JQP1 A0A087T137 A0A1Y1MSE2 X1WKK7 A0A1B6DER6 A0A0V1CRV4 A0A1Y1K4Z8 X1X571 A0A0V1CUA7 X1WSB4 A0A1B6E2X5 J9M3V0 X1WQE2 A0A0V1NJ43 A0A1B6MTB6 A0A087UAY4 A0A1B6LLL4 J9JUF3 Q207U0 A0A1B6KGD1 A0A1B6M803 E7BXG0 X1WSQ3 A0A1B6MIG5 A0A1B6DJR9 A0A0V1N525 A0A1B6MBV4 A0A023F1Y2 A0A0V1LC53 A0A2P8ZLW3 A0A087UHP8 A0A2P8XIJ0 A0A1Y1LGT7 A0A1B6LVC4 A0A0V0W8G5 A0A087TGK5 A0A1W4WUL1 A0A023F5Y4 A0A0K2TIZ0 A0A0V0SH12 A0A1B6CEU0 A0A1B6MC64 J9LFP4 A0A2P8Z261 A0A2P8YU38 A0A2H1V9L6 A0A023F5Y0 B0XHV3 A0A2S2R2M4 A0A2A4K028 A0A087U919 A0A087TH18 X1X9R9 J9JNY3 A0A1B6LUY7 A0A2R7WGP5 A0A146KVM3 A0A2P8XRS2 A0A3L8DT18 A0A1B6C4A1 A0A1B6CPZ3 D2XZW7 B1P5D5
A0A2A4J2P3 J9JSM6 A0A1B6BXA3 A0A1B6E438 A0A1B6JQP1 A0A087T137 A0A1Y1MSE2 X1WKK7 A0A1B6DER6 A0A0V1CRV4 A0A1Y1K4Z8 X1X571 A0A0V1CUA7 X1WSB4 A0A1B6E2X5 J9M3V0 X1WQE2 A0A0V1NJ43 A0A1B6MTB6 A0A087UAY4 A0A1B6LLL4 J9JUF3 Q207U0 A0A1B6KGD1 A0A1B6M803 E7BXG0 X1WSQ3 A0A1B6MIG5 A0A1B6DJR9 A0A0V1N525 A0A1B6MBV4 A0A023F1Y2 A0A0V1LC53 A0A2P8ZLW3 A0A087UHP8 A0A2P8XIJ0 A0A1Y1LGT7 A0A1B6LVC4 A0A0V0W8G5 A0A087TGK5 A0A1W4WUL1 A0A023F5Y4 A0A0K2TIZ0 A0A0V0SH12 A0A1B6CEU0 A0A1B6MC64 J9LFP4 A0A2P8Z261 A0A2P8YU38 A0A2H1V9L6 A0A023F5Y0 B0XHV3 A0A2S2R2M4 A0A2A4K028 A0A087U919 A0A087TH18 X1X9R9 J9JNY3 A0A1B6LUY7 A0A2R7WGP5 A0A146KVM3 A0A2P8XRS2 A0A3L8DT18 A0A1B6C4A1 A0A1B6CPZ3 D2XZW7 B1P5D5
Ontologies
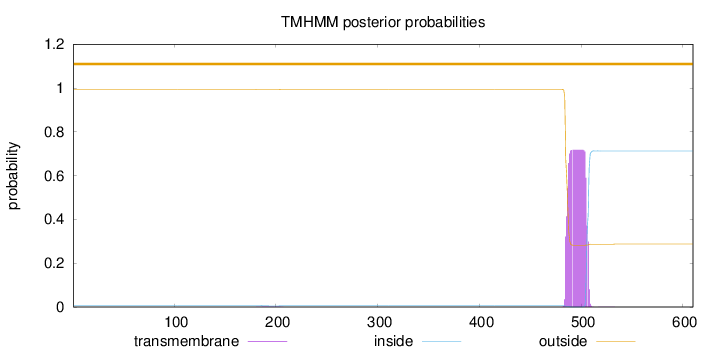
Topology
Length:
610
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.83214
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00695
outside
1 - 610
Population Genetic Test Statistics
Pi
267.271589
Theta
196.341126
Tajima's D
1.095786
CLR
0.197731
CSRT
0.683465826708665
Interpretation
Uncertain
