Gene
KWMTBOMO13324
Pre Gene Modal
BGIBMGA004678
Annotation
cytochrome_P450_CYP341B2_[Helicoverpa_armigera]
Location in the cell
Extracellular Reliability : 1.297 Mitochondrial Reliability : 1.268
Sequence
CDS
ATGAAAACGTTTCAACGTCTCGGCAAAGAGTCTACGGAGCGCGGAGGCTTATTGGCTCTCTGGCAGGGTAGCCGACTTTACGTCATGATATCAGAACCTGTTATGGCGGAGTACGTCCTCAAAACTTGTTTAGAAAAAGACGATATCTTGAAATGTTCCCGGTTCCTCGTCGGAAACGGAAGCGTGTTCGCTCCAGTTTCAATATGGCGTCCTCGTCGTAAAATCTTAGCTCCGACCTTCAGTCCTAAGAACCTAACACACTTTGTGGACATCTTCTCAAAACAAAGTTCTTATATGGTCAAATATCTTGGGAAAGCTGCAAAGACAGGGAATTTTTCAATATGGAAATACATAAATACATATTCTATGGATTCGATATGTGAAACGACATTAGGAGTGAAGGTGAATGCTCAAGGTAACTCCGAGCAACCTTTCCTGAGGGCCTTCGAAATAATTTGTCGTTTGGACTCATCTCGTTTCTGTCAGCCGTGGCTTCATAACGACACAGTGTACAAAATGATGCCACAGTACCAGCAGCATAAGGATTCCAAAGATTTTCTTTGTAATTTCATTGACCAGGTGATAAAATCAAAACGAAATAGTTTGGAGGAACAAAAGGATAGCACAGAAGCAGATCAAAATGCACATAGAAACGGCCTGAAATCATTTCTGGAATTGTTAATCGAGTCGTCAGGAGGGAATAAAGGCTACACTGATTTGGAACTTCAGGAGGAAACTCTAGTGCTTGTTCTGGCTGGTACTGACACATCAGCGGTTGGAGTAGCTTTTACATCTGTCATGCTTTCCAGGCATCAGGACGTGCAGGAGAAGGTTTACGAAGAATTGAAAGAAGTCTTTGGTGATTCCGATAGACCTATAGTTGCAGATGATCTGCCCAAATTGAAATATTTGGAGGCAGTGATAAAAGAAACAATGAGATTGTATCCCCCAGTGCCTCTCATCGTCAGGAAGGTCGATAAGGATGTAACGCTGCCAACAGGATTAACTCTGGTCAAAAACTGCGGTATAGTGATCAACATCTGGGCTGTGCACAGGAACCCGCTCTACTGGGGTGATGATGCCGATATATTTAGACCGGAACGTTTTATTGACACGCCCATAAAGCACCCCGCTGCGTTCATGGCCTTCAGTCATGGCCCTAGAGCGTGTATTGGTTATCAATACGCGACGATGTCAATGAAGACGGCAACAGCCAATCTCCTACGTCATTTCCGTCTCCGCCCAGCCGAGCCGACGGATCCAACATATAAACACGAAAAAAACAAGCCACTTCGCGTCAAATTCGATGTCATGATGAAAGATATGGACAATTTTACAGTGCAATTAGAACCTAGATACAAATAG
Protein
MKTFQRLGKESTERGGLLALWQGSRLYVMISEPVMAEYVLKTCLEKDDILKCSRFLVGNGSVFAPVSIWRPRRKILAPTFSPKNLTHFVDIFSKQSSYMVKYLGKAAKTGNFSIWKYINTYSMDSICETTLGVKVNAQGNSEQPFLRAFEIICRLDSSRFCQPWLHNDTVYKMMPQYQQHKDSKDFLCNFIDQVIKSKRNSLEEQKDSTEADQNAHRNGLKSFLELLIESSGGNKGYTDLELQEETLVLVLAGTDTSAVGVAFTSVMLSRHQDVQEKVYEELKEVFGDSDRPIVADDLPKLKYLEAVIKETMRLYPPVPLIVRKVDKDVTLPTGLTLVKNCGIVINIWAVHRNPLYWGDDADIFRPERFIDTPIKHPAAFMAFSHGPRACIGYQYATMSMKTATANLLRHFRLRPAEPTDPTYKHEKNKPLRVKFDVMMKDMDNFTVQLEPRYK
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
L0N785
H9J588
A0A068EVJ0
A0A286QUH1
A0A2A4JGQ9
A0A2A4ISM9
+ More
A0A2A4JHV7 A0A0K8TUL8 A0A0K8TUI4 A0A3S2PCK4 A0A2H1WBM8 A0A0D6A1C6 A0A068EU38 A0A2Z5SAV1 A0A2H1WBK5 A0A2H1VMK4 A0A248QED4 J7FJY6 A0A1E1WU74 A0A2A4IT41 A0A194PYM9 A0A2H1WUL9 A0A0K8TVP5 A0A194RF00 A0A0K8TV31 A0A248QEI3 A0A346IHZ3 A0A0K8TUH6 A0A194R0G6 S4VGT4 A0A2H1WUI5 A0A194Q3L1 J7FIT6 X5DB16 A0A346II01 A0A1V0D9H7 Q4R1I7 A0A3S2TR28 A0A194R208 A0A346IHY9 A0A0L7KRF5 A0A2A4IYA1 A0A2H1VP75 A0A194R193 A0A194REI2 A0A194R5Z5 A0A194PZG6 A0A1V0D9I5 A0A0L7L4L4 A0A0L7KSI3 A0A0C5C579 A0A212EGV4 A0A3S2M7I8 A0A194PY94 A0A2W1C3W1 A0A286MXL8 H9J5C7 A0A2A4JY96 A0A2H1V4V6 A0A2J7RF67 A0A068EU45 A0A2W1BZ42 A0A212FEU5 A0A1V0D9C8 A0A0C9RFM1 A0A0U3YU82 A0A2H1V533 A0A2J7QEQ2 A0A1A9W296 A0A194RMW0 A0A1W6L1D9 A0A194Q0P4 A0A2W1BKF4 Q16L54 A0A2J7QZP4 A0A2P8Z9S5 E9FYP2 Q16L49 A0A067RPH3 A0A1S2ZU39 A0A2H8U0V3 A0A1L8E485 A0A1S4FX34 A0A0C5C571 A0A3R7MJW5 I4DNC9 A0A1B0AJY5 J9JU30 A0A1L8E445 A0A0K8TV47 A0A067R8S0
A0A2A4JHV7 A0A0K8TUL8 A0A0K8TUI4 A0A3S2PCK4 A0A2H1WBM8 A0A0D6A1C6 A0A068EU38 A0A2Z5SAV1 A0A2H1WBK5 A0A2H1VMK4 A0A248QED4 J7FJY6 A0A1E1WU74 A0A2A4IT41 A0A194PYM9 A0A2H1WUL9 A0A0K8TVP5 A0A194RF00 A0A0K8TV31 A0A248QEI3 A0A346IHZ3 A0A0K8TUH6 A0A194R0G6 S4VGT4 A0A2H1WUI5 A0A194Q3L1 J7FIT6 X5DB16 A0A346II01 A0A1V0D9H7 Q4R1I7 A0A3S2TR28 A0A194R208 A0A346IHY9 A0A0L7KRF5 A0A2A4IYA1 A0A2H1VP75 A0A194R193 A0A194REI2 A0A194R5Z5 A0A194PZG6 A0A1V0D9I5 A0A0L7L4L4 A0A0L7KSI3 A0A0C5C579 A0A212EGV4 A0A3S2M7I8 A0A194PY94 A0A2W1C3W1 A0A286MXL8 H9J5C7 A0A2A4JY96 A0A2H1V4V6 A0A2J7RF67 A0A068EU45 A0A2W1BZ42 A0A212FEU5 A0A1V0D9C8 A0A0C9RFM1 A0A0U3YU82 A0A2H1V533 A0A2J7QEQ2 A0A1A9W296 A0A194RMW0 A0A1W6L1D9 A0A194Q0P4 A0A2W1BKF4 Q16L54 A0A2J7QZP4 A0A2P8Z9S5 E9FYP2 Q16L49 A0A067RPH3 A0A1S2ZU39 A0A2H8U0V3 A0A1L8E485 A0A1S4FX34 A0A0C5C571 A0A3R7MJW5 I4DNC9 A0A1B0AJY5 J9JU30 A0A1L8E445 A0A0K8TV47 A0A067R8S0
Pubmed
EMBL
AB436841
BAM73795.1
BABH01039691
KM016716
AID54868.1
KX443471
+ More
ASO98046.1 NWSH01001465 PCG71161.1 NWSH01008140 PCG62679.1 PCG71164.1 GCVX01000053 JAI18177.1 GCVX01000088 JAI18142.1 RSAL01000100 RVE47555.1 ODYU01007567 SOQ50447.1 AB795798 BAQ56325.1 KM016717 AID54869.1 LC326250 BBA58211.1 SOQ50448.1 ODYU01003376 SOQ42053.1 KX425016 ASN63843.1 JX310095 AFP20606.1 GDQN01000461 JAT90593.1 NWSH01007729 PCG62839.1 KQ459586 KPI98133.1 ODYU01011123 SOQ56666.1 GCVX01000090 JAI18140.1 KQ460323 KPJ15875.1 GCVX01000087 JAI18143.1 KX443470 ASO98045.1 MH138207 AXP17148.1 GCVX01000086 JAI18144.1 KQ460883 KPJ11283.1 KC789756 AGO62011.1 SOQ56667.1 KPI98000.1 JX310094 AFP20605.1 KF701170 AHW57340.1 MH138215 AXP17156.1 KY212088 ARA91652.1 AB201381 BAD99563.1 RSAL01000018 RVE52820.1 KPJ11285.1 MH138203 AXP17144.1 JTDY01006839 KOB65655.1 NWSH01005164 PCG64388.1 SOQ42054.1 KPJ11284.1 KPJ15874.1 KPJ11286.1 KPI98134.1 KY212089 ARA91653.1 JTDY01002945 KOB70438.1 JTDY01006414 KOB66031.1 KP001145 AJN91190.1 AGBW02015005 OWR40717.1 RVE52821.1 KPI97998.1 KZ149894 PZC78853.1 MF684336 ASX93970.1 BABH01037061 NWSH01000444 PCG76362.1 ODYU01000688 SOQ35885.1 NEVH01004413 PNF39482.1 KM016722 AID54874.1 PZC78854.1 AGBW02008898 OWR52238.1 KY212040 ARA91604.1 GBYB01007184 GBYB01007185 JAG76951.1 JAG76952.1 KT071008 ALX81393.1 SOQ35886.1 NEVH01015306 PNF27066.1 KQ459989 KPJ18754.1 KX609528 ARN17935.1 KQ459591 KPI96980.1 KZ150119 PZC73210.1 CH477923 EAT35034.2 NEVH01009068 PNF34047.1 PYGN01000135 PSN53242.1 GL732527 EFX87732.1 EAT35039.1 KK852503 KDR22530.1 GFXV01008004 MBW19809.1 GFDF01000640 JAV13444.1 KP001140 AJN91185.1 QCYY01003404 ROT63648.1 AK402852 BAM19419.1 ABLF02021072 ABLF02036370 ABLF02036380 GFDF01000581 JAV13503.1 GCVX01000094 JAI18136.1 KK852621 KDR20032.1
ASO98046.1 NWSH01001465 PCG71161.1 NWSH01008140 PCG62679.1 PCG71164.1 GCVX01000053 JAI18177.1 GCVX01000088 JAI18142.1 RSAL01000100 RVE47555.1 ODYU01007567 SOQ50447.1 AB795798 BAQ56325.1 KM016717 AID54869.1 LC326250 BBA58211.1 SOQ50448.1 ODYU01003376 SOQ42053.1 KX425016 ASN63843.1 JX310095 AFP20606.1 GDQN01000461 JAT90593.1 NWSH01007729 PCG62839.1 KQ459586 KPI98133.1 ODYU01011123 SOQ56666.1 GCVX01000090 JAI18140.1 KQ460323 KPJ15875.1 GCVX01000087 JAI18143.1 KX443470 ASO98045.1 MH138207 AXP17148.1 GCVX01000086 JAI18144.1 KQ460883 KPJ11283.1 KC789756 AGO62011.1 SOQ56667.1 KPI98000.1 JX310094 AFP20605.1 KF701170 AHW57340.1 MH138215 AXP17156.1 KY212088 ARA91652.1 AB201381 BAD99563.1 RSAL01000018 RVE52820.1 KPJ11285.1 MH138203 AXP17144.1 JTDY01006839 KOB65655.1 NWSH01005164 PCG64388.1 SOQ42054.1 KPJ11284.1 KPJ15874.1 KPJ11286.1 KPI98134.1 KY212089 ARA91653.1 JTDY01002945 KOB70438.1 JTDY01006414 KOB66031.1 KP001145 AJN91190.1 AGBW02015005 OWR40717.1 RVE52821.1 KPI97998.1 KZ149894 PZC78853.1 MF684336 ASX93970.1 BABH01037061 NWSH01000444 PCG76362.1 ODYU01000688 SOQ35885.1 NEVH01004413 PNF39482.1 KM016722 AID54874.1 PZC78854.1 AGBW02008898 OWR52238.1 KY212040 ARA91604.1 GBYB01007184 GBYB01007185 JAG76951.1 JAG76952.1 KT071008 ALX81393.1 SOQ35886.1 NEVH01015306 PNF27066.1 KQ459989 KPJ18754.1 KX609528 ARN17935.1 KQ459591 KPI96980.1 KZ150119 PZC73210.1 CH477923 EAT35034.2 NEVH01009068 PNF34047.1 PYGN01000135 PSN53242.1 GL732527 EFX87732.1 EAT35039.1 KK852503 KDR22530.1 GFXV01008004 MBW19809.1 GFDF01000640 JAV13444.1 KP001140 AJN91185.1 QCYY01003404 ROT63648.1 AK402852 BAM19419.1 ABLF02021072 ABLF02036370 ABLF02036380 GFDF01000581 JAV13503.1 GCVX01000094 JAI18136.1 KK852621 KDR20032.1
Proteomes
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
L0N785
H9J588
A0A068EVJ0
A0A286QUH1
A0A2A4JGQ9
A0A2A4ISM9
+ More
A0A2A4JHV7 A0A0K8TUL8 A0A0K8TUI4 A0A3S2PCK4 A0A2H1WBM8 A0A0D6A1C6 A0A068EU38 A0A2Z5SAV1 A0A2H1WBK5 A0A2H1VMK4 A0A248QED4 J7FJY6 A0A1E1WU74 A0A2A4IT41 A0A194PYM9 A0A2H1WUL9 A0A0K8TVP5 A0A194RF00 A0A0K8TV31 A0A248QEI3 A0A346IHZ3 A0A0K8TUH6 A0A194R0G6 S4VGT4 A0A2H1WUI5 A0A194Q3L1 J7FIT6 X5DB16 A0A346II01 A0A1V0D9H7 Q4R1I7 A0A3S2TR28 A0A194R208 A0A346IHY9 A0A0L7KRF5 A0A2A4IYA1 A0A2H1VP75 A0A194R193 A0A194REI2 A0A194R5Z5 A0A194PZG6 A0A1V0D9I5 A0A0L7L4L4 A0A0L7KSI3 A0A0C5C579 A0A212EGV4 A0A3S2M7I8 A0A194PY94 A0A2W1C3W1 A0A286MXL8 H9J5C7 A0A2A4JY96 A0A2H1V4V6 A0A2J7RF67 A0A068EU45 A0A2W1BZ42 A0A212FEU5 A0A1V0D9C8 A0A0C9RFM1 A0A0U3YU82 A0A2H1V533 A0A2J7QEQ2 A0A1A9W296 A0A194RMW0 A0A1W6L1D9 A0A194Q0P4 A0A2W1BKF4 Q16L54 A0A2J7QZP4 A0A2P8Z9S5 E9FYP2 Q16L49 A0A067RPH3 A0A1S2ZU39 A0A2H8U0V3 A0A1L8E485 A0A1S4FX34 A0A0C5C571 A0A3R7MJW5 I4DNC9 A0A1B0AJY5 J9JU30 A0A1L8E445 A0A0K8TV47 A0A067R8S0
A0A2A4JHV7 A0A0K8TUL8 A0A0K8TUI4 A0A3S2PCK4 A0A2H1WBM8 A0A0D6A1C6 A0A068EU38 A0A2Z5SAV1 A0A2H1WBK5 A0A2H1VMK4 A0A248QED4 J7FJY6 A0A1E1WU74 A0A2A4IT41 A0A194PYM9 A0A2H1WUL9 A0A0K8TVP5 A0A194RF00 A0A0K8TV31 A0A248QEI3 A0A346IHZ3 A0A0K8TUH6 A0A194R0G6 S4VGT4 A0A2H1WUI5 A0A194Q3L1 J7FIT6 X5DB16 A0A346II01 A0A1V0D9H7 Q4R1I7 A0A3S2TR28 A0A194R208 A0A346IHY9 A0A0L7KRF5 A0A2A4IYA1 A0A2H1VP75 A0A194R193 A0A194REI2 A0A194R5Z5 A0A194PZG6 A0A1V0D9I5 A0A0L7L4L4 A0A0L7KSI3 A0A0C5C579 A0A212EGV4 A0A3S2M7I8 A0A194PY94 A0A2W1C3W1 A0A286MXL8 H9J5C7 A0A2A4JY96 A0A2H1V4V6 A0A2J7RF67 A0A068EU45 A0A2W1BZ42 A0A212FEU5 A0A1V0D9C8 A0A0C9RFM1 A0A0U3YU82 A0A2H1V533 A0A2J7QEQ2 A0A1A9W296 A0A194RMW0 A0A1W6L1D9 A0A194Q0P4 A0A2W1BKF4 Q16L54 A0A2J7QZP4 A0A2P8Z9S5 E9FYP2 Q16L49 A0A067RPH3 A0A1S2ZU39 A0A2H8U0V3 A0A1L8E485 A0A1S4FX34 A0A0C5C571 A0A3R7MJW5 I4DNC9 A0A1B0AJY5 J9JU30 A0A1L8E445 A0A0K8TV47 A0A067R8S0
PDB
4D75
E-value=3.05892e-39,
Score=407
Ontologies
GO
Topology
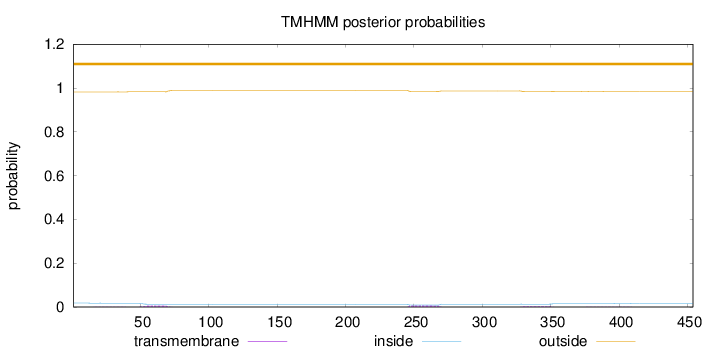
Length:
454
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.36743
Exp number, first 60 AAs:
0.05931
Total prob of N-in:
0.01779
outside
1 - 454
Population Genetic Test Statistics
Pi
197.594162
Theta
157.85275
Tajima's D
0.610767
CLR
0.110454
CSRT
0.545622718864057
Interpretation
Uncertain
