Gene
KWMTBOMO13323
Pre Gene Modal
BGIBMGA004677
Annotation
PREDICTED:_uncharacterized_protein_LOC105841761_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.77
Sequence
CDS
ATGCTGACGACGGTCCTGAACGACAGTCCTATTTTTCCCGAAAACGAACAGGAGCAAGCAGATTTATCGGCGCCATTACTCATGGACAGTCCGCAGTGCGTTTTATTCGGTACATTCATGATTTGGTTCGCATCAATAACTATTAATCTCGGCCCGACATTCCTGTCCGGGGCCCTGGCGGCCAGTGCCGGGTCTTATGGCTCGTACGGGCCGGCACCTTCGTGTCCCTTGGTGCGGGGTCCTTTCCGACATTACGTACTTAACGCGTTATGGATAGGCGTGAACGCTGTCTGCGTCGGGTTGACGCTTTTCCACTTGAGGAAACTCCACAGGGATCTCACTAAGGCTAACGTAGAGGCAGTTCGAGTAGCTGGTTTAGTGACAACGCTTGTCAGCATGGCAGGAGACAACCGGCGTATGGACTCTCTGCCCAACAGCGTTGGACCGAGGACTGTTGATGGTAACTTTTCTGATGGCCACTGCCGACCATCAGGCAGTACCAGTCTGCCTCTCGGCGGAGGGCGTCGTCTTCGAGGGTACTTGGCACGTGTTGAGCGAGAAGGAGTCAGACGAGTCCGCATGTTTCTTGTCATCACCGCGGCCTACGTACTCTTCTGGGGTCCATTGTTCATGATTACGCTGCTCCACCATCCAGCTCTGACATCACATGTGGCATATGAACTCTACTTCCAAATCACGTTGCACATTTCGTACGTGCACGCGGCGGTGAACCCGCTGTTGTTCATGGCGTTGCATCGTGCGCTGCGCCGCGCCGCCCTCCAGCTGTGCTGCGGCTGCTGTGTGCATTGGAGTCAATTCGTTCTAGCCCTTACTGCTGCAGGTAATGTATTAAAATCGAACGTTCTCGGACTGTGA
Protein
MLTTVLNDSPIFPENEQEQADLSAPLLMDSPQCVLFGTFMIWFASITINLGPTFLSGALAASAGSYGSYGPAPSCPLVRGPFRHYVLNALWIGVNAVCVGLTLFHLRKLHRDLTKANVEAVRVAGLVTTLVSMAGDNRRMDSLPNSVGPRTVDGNFSDGHCRPSGSTSLPLGGGRRLRGYLARVEREGVRRVRMFLVITAAYVLFWGPLFMITLLHHPALTSHVAYELYFQITLHISYVHAAVNPLLFMALHRALRRAALQLCCGCCVHWSQFVLALTAAGNVLKSNVLGL
Summary
Uniprot
H9J587
A0A2W1BRN8
A0A194PXH9
A0A194R0C7
A0A2A4J4D9
A0A2H1VRN6
+ More
A0A0L7LLY9 F4X170 A0A151XH50 A0A0C9QLH9 A0A0C9R8I4 A0A195DTS9 E9IJJ5 A0A195ETB4 A0A195AYH4 A0A067R438 A0A3L8DVL3 A0A026X2Y3 E2BLD7 E1ZWQ9 A0A2P8YEB3 A0A087ZT25 K7JLA5 A0A232F424 D7EJ14 T1HBM0 A0A2J7RJK3 J9KWM2 A0A2A3E786 A0A154P5J7 A0A151IMV9 T1IM87 A0A0K2TYB2 A0A0K2TYD1 A0A0K2TZ94 A0A0K2U047 A0A0L7RDY8 A0A2P6LE72 A0A2P6KVA2 B7PT77 A0A0P5NTZ8 A0A164Y831 A0A0P5WT20 A0A0P5ILY4 A0A0P6DVN2 A0A0P5KU74 A0A0P5L3K1 A0A0P6E7Z5 A0A0P6F5B2 A0A0P4XBM5 A0A0P5C9Z0 A0A0P5VE21 A0A0P5WCL4 A0A0P5UPX0 A0A0P5FKE7 A0A0P5AQA6 A0A0P5UU44 A0A267GGH6 A0A267DFW3 A0A267DD94 A0A267FC55
A0A0L7LLY9 F4X170 A0A151XH50 A0A0C9QLH9 A0A0C9R8I4 A0A195DTS9 E9IJJ5 A0A195ETB4 A0A195AYH4 A0A067R438 A0A3L8DVL3 A0A026X2Y3 E2BLD7 E1ZWQ9 A0A2P8YEB3 A0A087ZT25 K7JLA5 A0A232F424 D7EJ14 T1HBM0 A0A2J7RJK3 J9KWM2 A0A2A3E786 A0A154P5J7 A0A151IMV9 T1IM87 A0A0K2TYB2 A0A0K2TYD1 A0A0K2TZ94 A0A0K2U047 A0A0L7RDY8 A0A2P6LE72 A0A2P6KVA2 B7PT77 A0A0P5NTZ8 A0A164Y831 A0A0P5WT20 A0A0P5ILY4 A0A0P6DVN2 A0A0P5KU74 A0A0P5L3K1 A0A0P6E7Z5 A0A0P6F5B2 A0A0P4XBM5 A0A0P5C9Z0 A0A0P5VE21 A0A0P5WCL4 A0A0P5UPX0 A0A0P5FKE7 A0A0P5AQA6 A0A0P5UU44 A0A267GGH6 A0A267DFW3 A0A267DD94 A0A267FC55
Pubmed
EMBL
BABH01039701
KZ149981
PZC75817.1
KQ459586
KPI98051.1
KQ460883
+ More
KPJ11243.1 NWSH01003457 PCG66262.1 ODYU01004002 SOQ43436.1 JTDY01000641 KOB76369.1 GL888527 EGI59806.1 KQ982138 KYQ59645.1 GBYB01004394 JAG74161.1 GBYB01004395 JAG74162.1 KQ980390 KYN16283.1 GL763868 EFZ19236.1 KQ981989 KYN31152.1 KQ976711 KYM77014.1 KK852716 KDR17836.1 QOIP01000004 RLU24273.1 KK107046 EZA61759.1 GL449017 EFN83489.1 GL434886 EFN74387.1 PYGN01000665 PSN42588.1 NNAY01001029 OXU25425.1 KQ971326 EFA12490.2 ACPB03008540 NEVH01002992 PNF41012.1 ABLF02027042 ABLF02027043 ABLF02027044 ABLF02027046 ABLF02044812 KZ288349 PBC27364.1 KQ434823 KZC07215.1 KQ977047 KYN06106.1 JH430992 HACA01013648 CDW31009.1 HACA01013647 CDW31008.1 HACA01013646 CDW31007.1 HACA01013645 CDW31006.1 KQ414612 KOC69197.1 MWRG01000179 PRD36873.1 MWRG01004773 PRD30248.1 ABJB010061310 DS784171 EEC09799.1 GDIQ01137783 JAL13943.1 LRGB01000930 KZS14975.1 GDIP01082955 JAM20760.1 GDIQ01210844 JAK40881.1 GDIQ01075510 JAN19227.1 GDIQ01179465 JAK72260.1 GDIQ01180861 JAK70864.1 GDIQ01066597 JAN28140.1 GDIQ01065247 JAN29490.1 GDIP01246556 JAI76845.1 GDIP01173512 JAJ49890.1 GDIP01101096 JAM02619.1 GDIP01087986 JAM15729.1 GDIP01111888 JAL91826.1 GDIQ01255770 JAJ95954.1 GDIP01195567 JAJ27835.1 GDIP01125385 JAL78329.1 NIVC01000349 PAA85096.1 NIVC01004443 PAA47462.1 NIVC01004808 PAA46549.1 NIVC01001167 PAA71361.1
KPJ11243.1 NWSH01003457 PCG66262.1 ODYU01004002 SOQ43436.1 JTDY01000641 KOB76369.1 GL888527 EGI59806.1 KQ982138 KYQ59645.1 GBYB01004394 JAG74161.1 GBYB01004395 JAG74162.1 KQ980390 KYN16283.1 GL763868 EFZ19236.1 KQ981989 KYN31152.1 KQ976711 KYM77014.1 KK852716 KDR17836.1 QOIP01000004 RLU24273.1 KK107046 EZA61759.1 GL449017 EFN83489.1 GL434886 EFN74387.1 PYGN01000665 PSN42588.1 NNAY01001029 OXU25425.1 KQ971326 EFA12490.2 ACPB03008540 NEVH01002992 PNF41012.1 ABLF02027042 ABLF02027043 ABLF02027044 ABLF02027046 ABLF02044812 KZ288349 PBC27364.1 KQ434823 KZC07215.1 KQ977047 KYN06106.1 JH430992 HACA01013648 CDW31009.1 HACA01013647 CDW31008.1 HACA01013646 CDW31007.1 HACA01013645 CDW31006.1 KQ414612 KOC69197.1 MWRG01000179 PRD36873.1 MWRG01004773 PRD30248.1 ABJB010061310 DS784171 EEC09799.1 GDIQ01137783 JAL13943.1 LRGB01000930 KZS14975.1 GDIP01082955 JAM20760.1 GDIQ01210844 JAK40881.1 GDIQ01075510 JAN19227.1 GDIQ01179465 JAK72260.1 GDIQ01180861 JAK70864.1 GDIQ01066597 JAN28140.1 GDIQ01065247 JAN29490.1 GDIP01246556 JAI76845.1 GDIP01173512 JAJ49890.1 GDIP01101096 JAM02619.1 GDIP01087986 JAM15729.1 GDIP01111888 JAL91826.1 GDIQ01255770 JAJ95954.1 GDIP01195567 JAJ27835.1 GDIP01125385 JAL78329.1 NIVC01000349 PAA85096.1 NIVC01004443 PAA47462.1 NIVC01004808 PAA46549.1 NIVC01001167 PAA71361.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000037510
UP000007755
+ More
UP000075809 UP000078492 UP000078541 UP000078540 UP000027135 UP000279307 UP000053097 UP000008237 UP000000311 UP000245037 UP000005203 UP000002358 UP000215335 UP000007266 UP000015103 UP000235965 UP000007819 UP000242457 UP000076502 UP000078542 UP000053825 UP000001555 UP000076858 UP000215902
UP000075809 UP000078492 UP000078541 UP000078540 UP000027135 UP000279307 UP000053097 UP000008237 UP000000311 UP000245037 UP000005203 UP000002358 UP000215335 UP000007266 UP000015103 UP000235965 UP000007819 UP000242457 UP000076502 UP000078542 UP000053825 UP000001555 UP000076858 UP000215902
PRIDE
Pfam
PF00001 7tm_1
ProteinModelPortal
H9J587
A0A2W1BRN8
A0A194PXH9
A0A194R0C7
A0A2A4J4D9
A0A2H1VRN6
+ More
A0A0L7LLY9 F4X170 A0A151XH50 A0A0C9QLH9 A0A0C9R8I4 A0A195DTS9 E9IJJ5 A0A195ETB4 A0A195AYH4 A0A067R438 A0A3L8DVL3 A0A026X2Y3 E2BLD7 E1ZWQ9 A0A2P8YEB3 A0A087ZT25 K7JLA5 A0A232F424 D7EJ14 T1HBM0 A0A2J7RJK3 J9KWM2 A0A2A3E786 A0A154P5J7 A0A151IMV9 T1IM87 A0A0K2TYB2 A0A0K2TYD1 A0A0K2TZ94 A0A0K2U047 A0A0L7RDY8 A0A2P6LE72 A0A2P6KVA2 B7PT77 A0A0P5NTZ8 A0A164Y831 A0A0P5WT20 A0A0P5ILY4 A0A0P6DVN2 A0A0P5KU74 A0A0P5L3K1 A0A0P6E7Z5 A0A0P6F5B2 A0A0P4XBM5 A0A0P5C9Z0 A0A0P5VE21 A0A0P5WCL4 A0A0P5UPX0 A0A0P5FKE7 A0A0P5AQA6 A0A0P5UU44 A0A267GGH6 A0A267DFW3 A0A267DD94 A0A267FC55
A0A0L7LLY9 F4X170 A0A151XH50 A0A0C9QLH9 A0A0C9R8I4 A0A195DTS9 E9IJJ5 A0A195ETB4 A0A195AYH4 A0A067R438 A0A3L8DVL3 A0A026X2Y3 E2BLD7 E1ZWQ9 A0A2P8YEB3 A0A087ZT25 K7JLA5 A0A232F424 D7EJ14 T1HBM0 A0A2J7RJK3 J9KWM2 A0A2A3E786 A0A154P5J7 A0A151IMV9 T1IM87 A0A0K2TYB2 A0A0K2TYD1 A0A0K2TZ94 A0A0K2U047 A0A0L7RDY8 A0A2P6LE72 A0A2P6KVA2 B7PT77 A0A0P5NTZ8 A0A164Y831 A0A0P5WT20 A0A0P5ILY4 A0A0P6DVN2 A0A0P5KU74 A0A0P5L3K1 A0A0P6E7Z5 A0A0P6F5B2 A0A0P4XBM5 A0A0P5C9Z0 A0A0P5VE21 A0A0P5WCL4 A0A0P5UPX0 A0A0P5FKE7 A0A0P5AQA6 A0A0P5UU44 A0A267GGH6 A0A267DFW3 A0A267DD94 A0A267FC55
PDB
3PBL
E-value=0.025936,
Score=86
Ontologies
GO
Topology
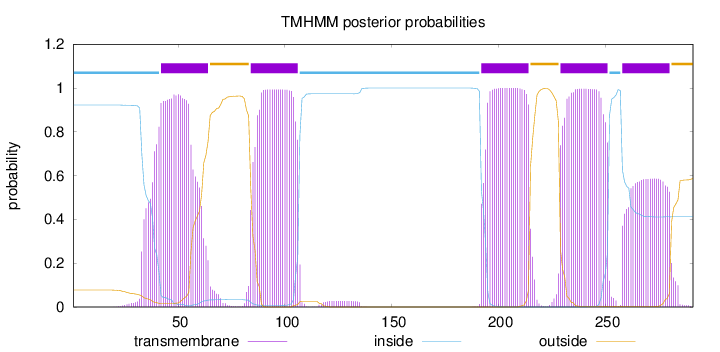
Length:
291
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
102.80279
Exp number, first 60 AAs:
21.47679
Total prob of N-in:
0.92257
POSSIBLE N-term signal
sequence
inside
1 - 41
TMhelix
42 - 64
outside
65 - 83
TMhelix
84 - 106
inside
107 - 191
TMhelix
192 - 214
outside
215 - 228
TMhelix
229 - 251
inside
252 - 257
TMhelix
258 - 280
outside
281 - 291
Population Genetic Test Statistics
Pi
40.020193
Theta
140.661944
Tajima's D
-0.91221
CLR
1.993473
CSRT
0.155092245387731
Interpretation
Uncertain
