Pre Gene Modal
BGIBMGA004687
Annotation
PREDICTED:_cyclin-G-associated_kinase_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.315
Sequence
CDS
ATGGGGTACTTCAGCTCAAGTGGCGCAAACGGTGGTAGTGAAAATGATTTTGTTGGTCAATTTGTCGAAATCGGTAACATGAAATTGAGAGTAAAAAAGGTGATTGCAGAAGGTGGTTTTGCATTTGTGTTTGTTGCGCAAGATGTAACTACTGGCACTGAATATGCCTTAAAGAGATTAATGGCTGCTGATGAACAAGCAAACAAGAACATCATCCAAGAGATCAGTATTCTAAAGAAGCTGTCAGGTCATCCAAATATAATTAAATACATAGCTGCATCGTTTATTGATAAAACAAAAACCTCTCATGGGATGGGAGAGTACCTTCTGCTGACTGATCTTTGCAGCGGTGGAAGTCTGATGGAGGCCTTACAGAATAGAGGGCAGGCATTCCCTCTACCAACAATACTTAGAGTATTTTATCAAACCTGTAGAGCAGTTCAGCACATGCACGCACAAGTCCCACCTATTGCGCATCGTGATCTTAAATTAGAAAATTTTCTAATATCCAATGAAGGAACAATGAAGTTGTGTGATTTCGGGTCTGCCACAACTGATGTGTACTCGCCTAATCCATCATGGAGTGCTAACCAAAGAAATATGTTGGAAGAAAATTTGGCCTTATTCACTACGCCTATGTACCGTGCTCCAGAGATGTTAGACACCTGGGATAACCGGAAAATAGACCACAGCGTCGATATGTGGGCTTTGGGGTGCATATTATACACCTTGTGTTACATGCAACACCCCTTCGAAGATTCCGCTAAGCTGGCAATACTTAATGGAAATTACAATTTGAACCCTAACGATCAACGTTACAAATGTTTTCATGAAATCATAAGTGGATGTCTAACAGTGAATCCCGCTGACAGGATGACGATAACCAATGTGTTGGAGCGGCTGGCTGCGATAGCGGAAACTCAGAATGTCAATCTCAAGCAACCACTGAAATTTGAGCGTAAAAAAGTTGAACAATCTGTGGCCACTAGCTCGCCAGGTGTATTCAACTTTTTCGGATGCCCCATAGTATTCTCAGATCCTGTATCAAACAGCCAGGAAGCACCGAATTCCAGGCCCCCGGAGCCCAGCCGCCCCCCGCCCCCCGCGCGGCCCCCGGCAGCGCCCTACCACCAGCCGCCGCTGCCGCAACCGCCCGTGAATTCTGGAGGACTCTTTTCCTCGATAAAAGGAGGAGCGGGAAGCTTCTTGAAGAATTTGAAAGACACTTCATCGAAGGTTATGCAAACTGTACAACAGTCTATTGCGAGAACCGATTTAGACATAAGCTACATTACCAGTCGTGTTATTGTAATGCCATATCCTTCTGAAGGCCTCGAGAGTGCATATAAGACTAATCACATCGATGATGTTAGGGCATTTCTGGAGAGTCGTCACCCTGGCGGGAAGTACTGCGTGTACAACGCGTGTCGCGGCGGCCGCCTCACGTTCGGGCGCCACGTGCTCGTGACGGACGCGTGCCGCGCGCTGTGGCCCACGGGCGCGCACCGAGCCCCGCTGTTGGCGCCCTACTACGCGCTGCTGCAGCACATGTACCAGTATCTAGCCAAGGACGACCGTAACGCCATCGTTATTACGTGTCAAGACGGCAAACAGATGTCGTGCATGTTGGTGTGCGGACTGCTGCTGTACTCGAAACTGGTGACCGTGCCGGAGGATGCGCTGCAGATATTCGCGGTCAAGAGATCCCCCATCAACATGCTGCCTAGTCAACTCAGATACTTATATATGCTTAGTAATATCATAAGACCAGATCCCATACTTCCTCACTTCAAGCCGGTGTCGTTAATATCTATGACGATACAGCCAGTTCCGCTGTTTACAAAAGCAAGGGACGGATGTCGACCGTACATTGAGATATACAATGAAGATCGTCTTGTTTTATCAACTCTACAAGATTACGATAGATTGCATCTCTATGGAATAGCCGAGGGAAAGGTAACGTTACCGTTGGACAGCACGGTGACTGGCGACGTGTGCATTTGTGTGCTACACGCGAGACAGCAGCTCGGCCGAGTGCTGGCCGTGCCCGTACTGTCCCTGCACTTCCACACCGGCGCCCTCTCGTACGATCAGCACGCTATTAAATTTACAAGATCCGAAATAGACGGTATCGATGAAACGGTCCCAGTGAACGATCATTTTTCGGCGAACACGTCCGTTATCATAAGTCTCGTGGTACAAAACGGCGAGAGGAGGAAACCTCGCAGCGACCTCTGGGAAGACGACCCGTCGTTTCCGGTTCGCACTCCGGACGTACTGTTCTCTACTACATTAGAGAGGGACGAAACTATCGATAACTTTGTAACCGCAGACTACCCTCCAAGAGAGGTAGAGAAGCCCAGGCCATCCCGTCCCGCCCCTCCCTCTCCGCAGCCCCCGCAGCGGCCCCCTCCCCCGCGGCCGCTCCCCGCAGAGCCACAGGAGAACACAGCAGACTTCTTGAACTTAAACACGAACCAAACTCGTCAATCGGAAGAGAAAACCGAGAAGAAATTGAAGAACGAAGATTCTTTCGACTTTTTTGGGATGATGGAGAAAGAGACTGATGATACATTTTGTGATTTCCTCAGCAACAACGCGGAGAAAGCGCAAGTATGCGATTTAACACTAATCATACCAACACTTACTGATACCTATTTTTACTATAGCGGGTAG
Protein
MGYFSSSGANGGSENDFVGQFVEIGNMKLRVKKVIAEGGFAFVFVAQDVTTGTEYALKRLMAADEQANKNIIQEISILKKLSGHPNIIKYIAASFIDKTKTSHGMGEYLLLTDLCSGGSLMEALQNRGQAFPLPTILRVFYQTCRAVQHMHAQVPPIAHRDLKLENFLISNEGTMKLCDFGSATTDVYSPNPSWSANQRNMLEENLALFTTPMYRAPEMLDTWDNRKIDHSVDMWALGCILYTLCYMQHPFEDSAKLAILNGNYNLNPNDQRYKCFHEIISGCLTVNPADRMTITNVLERLAAIAETQNVNLKQPLKFERKKVEQSVATSSPGVFNFFGCPIVFSDPVSNSQEAPNSRPPEPSRPPPPARPPAAPYHQPPLPQPPVNSGGLFSSIKGGAGSFLKNLKDTSSKVMQTVQQSIARTDLDISYITSRVIVMPYPSEGLESAYKTNHIDDVRAFLESRHPGGKYCVYNACRGGRLTFGRHVLVTDACRALWPTGAHRAPLLAPYYALLQHMYQYLAKDDRNAIVITCQDGKQMSCMLVCGLLLYSKLVTVPEDALQIFAVKRSPINMLPSQLRYLYMLSNIIRPDPILPHFKPVSLISMTIQPVPLFTKARDGCRPYIEIYNEDRLVLSTLQDYDRLHLYGIAEGKVTLPLDSTVTGDVCICVLHARQQLGRVLAVPVLSLHFHTGALSYDQHAIKFTRSEIDGIDETVPVNDHFSANTSVIISLVVQNGERRKPRSDLWEDDPSFPVRTPDVLFSTTLERDETIDNFVTADYPPREVEKPRPSRPAPPSPQPPQRPPPPRPLPAEPQENTADFLNLNTNQTRQSEEKTEKKLKNEDSFDFFGMMEKETDDTFCDFLSNNAEKAQVCDLTLIIPTLTDTYFYYSG
Summary
Uniprot
A0A194PXI4
A0A212FFD0
A0A194R160
D6X251
N6T6J0
U4U733
+ More
Q17A59 A0A336MHS1 A0A1I8MMF9 A0A0A1XKW7 A0A1I8PUS5 A0A1A9W7D6 W8BG22 A0A1A9VG08 A0A0L0BMS5 A0A3R7PUI8 A0A240SWM5 A0A1B0B2R3 A0A240SX85 A0A1B0GG95 A0A0K8VGN7 B4PTP0 Q9VMY8 A0A0B4KG14 B4K713 B4QUY1 B4I3J9 B3P266 Q295D0 A0A1W4VY37 A0A023F137 B4GM74 B3LYZ9 T1JLE5 A0A069DZQ8 A0A1B0GPQ5 B4MC64 B4JGR2 A0A0P5VNA8 A0A0P5Y6B9 A0A0P5TW80 A0A0P6A7E0 A0A0P5UDD7 A0A0P4XZ30 A0A0P4XST3 A0A164Q148 A0A0M5J946 A0A0P5RV93 A0A0P5S6T5 A0A0P6E617 A0A0N8DZ61 A0A0P5A9S0 B4N8J4 A0A0P5SL88 A0A2R5LDN2 A0A3S1A2E1 A0A131XEI8 A0A224YQV2 L7LWY0 A0A0A9W7M2 A0A2C9JQC1 A0A2C9JQD0 A0A3Q3LBB4 U3FX76 A0A2D4F1Q8 A0A2D4F1R6 U3JS24 A0A0K8RXN4 A0A3B5ATE3 A0A0F7ZB68 A0A3Q3Q7F8 H2L7B0 A0A3Q1BGV1 A0A3P9IBW7 A0A3B3HZU6 A0A3P9IBY7 A0A3B3HBB7 A0A3P9KHW4 A0A3B3DIZ1 A0A3B3C2K8 A0A3P8VF38 A0A3P8V930 A0A2I4AI59 A0A3P8QFX5 A0A2I4AHZ3 A0A3Q2VD12 A0A3P9C643 A0A3Q2E2Q8 A0A3P8QG33 A0A3P9C5E2 A0A3Q2VD35 A0A3P8QHH4 A0A3P9C5J9 A0A3Q2VES1
Q17A59 A0A336MHS1 A0A1I8MMF9 A0A0A1XKW7 A0A1I8PUS5 A0A1A9W7D6 W8BG22 A0A1A9VG08 A0A0L0BMS5 A0A3R7PUI8 A0A240SWM5 A0A1B0B2R3 A0A240SX85 A0A1B0GG95 A0A0K8VGN7 B4PTP0 Q9VMY8 A0A0B4KG14 B4K713 B4QUY1 B4I3J9 B3P266 Q295D0 A0A1W4VY37 A0A023F137 B4GM74 B3LYZ9 T1JLE5 A0A069DZQ8 A0A1B0GPQ5 B4MC64 B4JGR2 A0A0P5VNA8 A0A0P5Y6B9 A0A0P5TW80 A0A0P6A7E0 A0A0P5UDD7 A0A0P4XZ30 A0A0P4XST3 A0A164Q148 A0A0M5J946 A0A0P5RV93 A0A0P5S6T5 A0A0P6E617 A0A0N8DZ61 A0A0P5A9S0 B4N8J4 A0A0P5SL88 A0A2R5LDN2 A0A3S1A2E1 A0A131XEI8 A0A224YQV2 L7LWY0 A0A0A9W7M2 A0A2C9JQC1 A0A2C9JQD0 A0A3Q3LBB4 U3FX76 A0A2D4F1Q8 A0A2D4F1R6 U3JS24 A0A0K8RXN4 A0A3B5ATE3 A0A0F7ZB68 A0A3Q3Q7F8 H2L7B0 A0A3Q1BGV1 A0A3P9IBW7 A0A3B3HZU6 A0A3P9IBY7 A0A3B3HBB7 A0A3P9KHW4 A0A3B3DIZ1 A0A3B3C2K8 A0A3P8VF38 A0A3P8V930 A0A2I4AI59 A0A3P8QFX5 A0A2I4AHZ3 A0A3Q2VD12 A0A3P9C643 A0A3Q2E2Q8 A0A3P8QG33 A0A3P9C5E2 A0A3Q2VD35 A0A3P8QHH4 A0A3P9C5J9 A0A3Q2VES1
Pubmed
26354079
22118469
18362917
19820115
23537049
17510324
+ More
25315136 25830018 24495485 26108605 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25474469 26334808 28049606 28797301 25576852 25401762 15562597 23915248 25727380 17554307 29451363 24487278 25186727
25315136 25830018 24495485 26108605 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25474469 26334808 28049606 28797301 25576852 25401762 15562597 23915248 25727380 17554307 29451363 24487278 25186727
EMBL
KQ459586
KPI98056.1
AGBW02008829
OWR52439.1
KQ460883
KPJ11249.1
+ More
KQ971371 EFA09916.1 APGK01041757 KB740997 ENN75844.1 KB632100 ERL88847.1 CH477340 EAT43105.1 UFQT01000897 SSX27877.1 GBXI01002303 JAD11989.1 GAMC01014314 JAB92241.1 JRES01001623 KNC21390.1 QCYY01001521 ROT77449.1 JXJN01007687 CCAG010001986 GDHF01014263 JAI38051.1 CM000160 EDW95623.1 AE014297 BT031027 AAF52168.1 AAF52169.1 ABV82409.1 AGB95626.1 CH933806 EDW15300.1 CM000364 EDX11562.1 CH480821 EDW54792.1 CH954181 EDV47744.1 CM000070 EAL28782.2 GBBI01004008 JAC14704.1 CH479185 EDW37948.1 CH902617 EDV41873.1 JH431979 GBGD01000215 JAC88674.1 AJVK01059969 CH940656 EDW58685.1 CH916369 EDV92666.1 GDIP01097723 JAM05992.1 GDIP01062432 JAM41283.1 GDIP01125825 JAL77889.1 GDIP01034738 JAM68977.1 GDIP01114674 JAL89040.1 GDIP01234863 JAI88538.1 GDIP01237074 JAI86327.1 LRGB01002486 KZS07336.1 CP012526 ALC46161.1 GDIQ01096174 JAL55552.1 GDIQ01097980 JAL53746.1 GDIQ01067493 JAN27244.1 GDIQ01077580 JAN17157.1 GDIP01205514 JAJ17888.1 CH964232 EDW81445.2 GDIQ01094225 JAL57501.1 GGLE01003460 MBY07586.1 RQTK01000002 RUS92097.1 GEFH01004545 JAP64036.1 GFPF01006993 MAA18139.1 GACK01009675 JAA55359.1 GBHO01039815 GBHO01032519 GBHO01016949 GBHO01016947 JAG03789.1 JAG11085.1 JAG26655.1 JAG26657.1 GAEP01000400 JAB54421.1 IACJ01046139 LAA41404.1 IACJ01046140 IACJ01046144 LAA41405.1 AGTO01011095 GBKC01000860 JAG45210.1 GBEX01000844 JAI13716.1
KQ971371 EFA09916.1 APGK01041757 KB740997 ENN75844.1 KB632100 ERL88847.1 CH477340 EAT43105.1 UFQT01000897 SSX27877.1 GBXI01002303 JAD11989.1 GAMC01014314 JAB92241.1 JRES01001623 KNC21390.1 QCYY01001521 ROT77449.1 JXJN01007687 CCAG010001986 GDHF01014263 JAI38051.1 CM000160 EDW95623.1 AE014297 BT031027 AAF52168.1 AAF52169.1 ABV82409.1 AGB95626.1 CH933806 EDW15300.1 CM000364 EDX11562.1 CH480821 EDW54792.1 CH954181 EDV47744.1 CM000070 EAL28782.2 GBBI01004008 JAC14704.1 CH479185 EDW37948.1 CH902617 EDV41873.1 JH431979 GBGD01000215 JAC88674.1 AJVK01059969 CH940656 EDW58685.1 CH916369 EDV92666.1 GDIP01097723 JAM05992.1 GDIP01062432 JAM41283.1 GDIP01125825 JAL77889.1 GDIP01034738 JAM68977.1 GDIP01114674 JAL89040.1 GDIP01234863 JAI88538.1 GDIP01237074 JAI86327.1 LRGB01002486 KZS07336.1 CP012526 ALC46161.1 GDIQ01096174 JAL55552.1 GDIQ01097980 JAL53746.1 GDIQ01067493 JAN27244.1 GDIQ01077580 JAN17157.1 GDIP01205514 JAJ17888.1 CH964232 EDW81445.2 GDIQ01094225 JAL57501.1 GGLE01003460 MBY07586.1 RQTK01000002 RUS92097.1 GEFH01004545 JAP64036.1 GFPF01006993 MAA18139.1 GACK01009675 JAA55359.1 GBHO01039815 GBHO01032519 GBHO01016949 GBHO01016947 JAG03789.1 JAG11085.1 JAG26655.1 JAG26657.1 GAEP01000400 JAB54421.1 IACJ01046139 LAA41404.1 IACJ01046140 IACJ01046144 LAA41405.1 AGTO01011095 GBKC01000860 JAG45210.1 GBEX01000844 JAI13716.1
Proteomes
UP000053268
UP000007151
UP000053240
UP000007266
UP000019118
UP000030742
+ More
UP000008820 UP000095301 UP000095300 UP000091820 UP000078200 UP000037069 UP000283509 UP000092443 UP000092460 UP000092445 UP000092444 UP000002282 UP000000803 UP000009192 UP000000304 UP000001292 UP000008711 UP000001819 UP000192221 UP000008744 UP000007801 UP000092462 UP000008792 UP000001070 UP000076858 UP000092553 UP000007798 UP000271974 UP000076420 UP000261640 UP000016665 UP000261400 UP000261600 UP000001038 UP000257160 UP000265200 UP000265180 UP000261560 UP000265120 UP000192220 UP000265100 UP000264840 UP000265160 UP000265020
UP000008820 UP000095301 UP000095300 UP000091820 UP000078200 UP000037069 UP000283509 UP000092443 UP000092460 UP000092445 UP000092444 UP000002282 UP000000803 UP000009192 UP000000304 UP000001292 UP000008711 UP000001819 UP000192221 UP000008744 UP000007801 UP000092462 UP000008792 UP000001070 UP000076858 UP000092553 UP000007798 UP000271974 UP000076420 UP000261640 UP000016665 UP000261400 UP000261600 UP000001038 UP000257160 UP000265200 UP000265180 UP000261560 UP000265120 UP000192220 UP000265100 UP000264840 UP000265160 UP000265020
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A194PXI4
A0A212FFD0
A0A194R160
D6X251
N6T6J0
U4U733
+ More
Q17A59 A0A336MHS1 A0A1I8MMF9 A0A0A1XKW7 A0A1I8PUS5 A0A1A9W7D6 W8BG22 A0A1A9VG08 A0A0L0BMS5 A0A3R7PUI8 A0A240SWM5 A0A1B0B2R3 A0A240SX85 A0A1B0GG95 A0A0K8VGN7 B4PTP0 Q9VMY8 A0A0B4KG14 B4K713 B4QUY1 B4I3J9 B3P266 Q295D0 A0A1W4VY37 A0A023F137 B4GM74 B3LYZ9 T1JLE5 A0A069DZQ8 A0A1B0GPQ5 B4MC64 B4JGR2 A0A0P5VNA8 A0A0P5Y6B9 A0A0P5TW80 A0A0P6A7E0 A0A0P5UDD7 A0A0P4XZ30 A0A0P4XST3 A0A164Q148 A0A0M5J946 A0A0P5RV93 A0A0P5S6T5 A0A0P6E617 A0A0N8DZ61 A0A0P5A9S0 B4N8J4 A0A0P5SL88 A0A2R5LDN2 A0A3S1A2E1 A0A131XEI8 A0A224YQV2 L7LWY0 A0A0A9W7M2 A0A2C9JQC1 A0A2C9JQD0 A0A3Q3LBB4 U3FX76 A0A2D4F1Q8 A0A2D4F1R6 U3JS24 A0A0K8RXN4 A0A3B5ATE3 A0A0F7ZB68 A0A3Q3Q7F8 H2L7B0 A0A3Q1BGV1 A0A3P9IBW7 A0A3B3HZU6 A0A3P9IBY7 A0A3B3HBB7 A0A3P9KHW4 A0A3B3DIZ1 A0A3B3C2K8 A0A3P8VF38 A0A3P8V930 A0A2I4AI59 A0A3P8QFX5 A0A2I4AHZ3 A0A3Q2VD12 A0A3P9C643 A0A3Q2E2Q8 A0A3P8QG33 A0A3P9C5E2 A0A3Q2VD35 A0A3P8QHH4 A0A3P9C5J9 A0A3Q2VES1
Q17A59 A0A336MHS1 A0A1I8MMF9 A0A0A1XKW7 A0A1I8PUS5 A0A1A9W7D6 W8BG22 A0A1A9VG08 A0A0L0BMS5 A0A3R7PUI8 A0A240SWM5 A0A1B0B2R3 A0A240SX85 A0A1B0GG95 A0A0K8VGN7 B4PTP0 Q9VMY8 A0A0B4KG14 B4K713 B4QUY1 B4I3J9 B3P266 Q295D0 A0A1W4VY37 A0A023F137 B4GM74 B3LYZ9 T1JLE5 A0A069DZQ8 A0A1B0GPQ5 B4MC64 B4JGR2 A0A0P5VNA8 A0A0P5Y6B9 A0A0P5TW80 A0A0P6A7E0 A0A0P5UDD7 A0A0P4XZ30 A0A0P4XST3 A0A164Q148 A0A0M5J946 A0A0P5RV93 A0A0P5S6T5 A0A0P6E617 A0A0N8DZ61 A0A0P5A9S0 B4N8J4 A0A0P5SL88 A0A2R5LDN2 A0A3S1A2E1 A0A131XEI8 A0A224YQV2 L7LWY0 A0A0A9W7M2 A0A2C9JQC1 A0A2C9JQD0 A0A3Q3LBB4 U3FX76 A0A2D4F1Q8 A0A2D4F1R6 U3JS24 A0A0K8RXN4 A0A3B5ATE3 A0A0F7ZB68 A0A3Q3Q7F8 H2L7B0 A0A3Q1BGV1 A0A3P9IBW7 A0A3B3HZU6 A0A3P9IBY7 A0A3B3HBB7 A0A3P9KHW4 A0A3B3DIZ1 A0A3B3C2K8 A0A3P8VF38 A0A3P8V930 A0A2I4AI59 A0A3P8QFX5 A0A2I4AHZ3 A0A3Q2VD12 A0A3P9C643 A0A3Q2E2Q8 A0A3P8QG33 A0A3P9C5E2 A0A3Q2VD35 A0A3P8QHH4 A0A3P9C5J9 A0A3Q2VES1
PDB
4O38
E-value=1.38825e-102,
Score=956
Ontologies
KEGG
GO
GO:0005524
GO:0004672
GO:0072583
GO:0043231
GO:0030276
GO:0031982
GO:0005737
GO:0072318
GO:0006898
GO:1901215
GO:0007291
GO:0001745
GO:0005795
GO:0006468
GO:0016021
GO:0004674
GO:0005829
GO:0007030
GO:0005794
GO:0090160
GO:0072659
GO:0010977
GO:1905224
GO:0034067
GO:0048471
GO:0005515
GO:0008270
GO:0051287
GO:0048869
GO:0003676
GO:0016491
GO:0015930
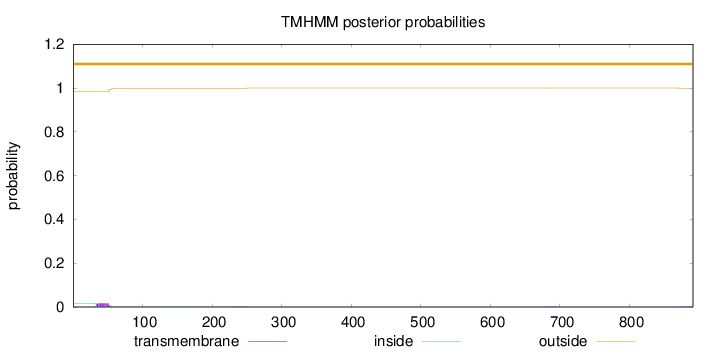
Topology
Length:
891
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.353400000000001
Exp number, first 60 AAs:
0.32871
Total prob of N-in:
0.01732
outside
1 - 891
Population Genetic Test Statistics
Pi
225.445417
Theta
166.989458
Tajima's D
0.461945
CLR
0.74687
CSRT
0.503424828758562
Interpretation
Uncertain
