Gene
KWMTBOMO13309
Pre Gene Modal
BGIBMGA004699
Annotation
PREDICTED:_kelch-like_ECH-associated_protein_1_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 1.982
Sequence
CDS
ATGGAAACTGACGAAGACTCGCGACGGACCAACATGCTGGATGACCTGCCTCCGATTAGTTGTGACAGCAACTTCGAGGATTCCGCCGGGAGTGGGTCCAACGAGATCGGCGATATGACATTCTGCATGGGAAATTACATCAGTGAGTTCATGAAGATGATGTTTACAATGAGATCACACCAAATGCTGACTGATGTCGTGTTGGAAGTTGGAAACGAACTCTTTCACGTCCACAAAGTTGTATTAGCGGCGGGCAGTCCTTATTTTAAGGCGATGTTCACGAGCGGCCTCAAGGAGTGTGAAATGTCAAGGGTCCGGCTACAAGGCGTGTGTCCTTCAGCGATGGCCTGGCTGATATACTTTATGTACACGGGGAAGATCAGGATAACTGAAGTGACGGTCTGCCAGCTGCTCCCGGCCGCCACCATGTTCCAGATAACGAATGTAATAAACGCATGCAGCGCGTTCCTCGAGCGACAGCTGGACCCCTCGAACGCGATCGGCATCGCGAACTTCGCCGAACAACACGGATGCGTTGAACTCAAGAAGAGAGCCAATCAATTTATCGAGAGACATTTCACGCAGGTGTGCCAAGAAGAAGAATTCCTACAACTAACAACACAAGAACTAATATCTCTTATCCGCAAAGACGAACTGAACGTAAGAGAAGAAAGAGAAGTGTATAACGCTGTTTTAAGTTGGGTAAAATACGATGAGGACAGAAGGCATCCCCGGATGGAGCACATCCTGCAGGCGGTCCGATGCCAGTACCTAACACCGAGCTTCCTGAAGGAGCAAATGAGCACGTGTACGGTTCTAAAGAAAGTACCTGCCTGTAGGGAATACTTAGCTAAGATATTCAAGGATCTGACATTACACAAGAACCCGGTCGTGAAAGAACGCAGACCGAACACGCCCCGCATTATATACGTGGCTGGCGGGTACTTCAGGCACTCGATAGACACGTTCGAGTCGTTCAACCTAGACGACAACTGCTGGACGACTCTGCCGCGACTGACCGTGCCCAGGTCGGGACTCGGAGCGGCGTTTTTAAAGGTAATACAGTGA
Protein
METDEDSRRTNMLDDLPPISCDSNFEDSAGSGSNEIGDMTFCMGNYISEFMKMMFTMRSHQMLTDVVLEVGNELFHVHKVVLAAGSPYFKAMFTSGLKECEMSRVRLQGVCPSAMAWLIYFMYTGKIRITEVTVCQLLPAATMFQITNVINACSAFLERQLDPSNAIGIANFAEQHGCVELKKRANQFIERHFTQVCQEEEFLQLTTQELISLIRKDELNVREEREVYNAVLSWVKYDEDRRHPRMEHILQAVRCQYLTPSFLKEQMSTCTVLKKVPACREYLAKIFKDLTLHKNPVVKERRPNTPRIIYVAGGYFRHSIDTFESFNLDDNCWTTLPRLTVPRSGLGAAFLKVIQ
Summary
Uniprot
A0A2P1JMP2
A0A2A4JEU7
A0A2A4JEC7
A0A2H4G7J0
A0A194PZ49
A0A194R1X7
+ More
A0A3S2NUV0 A0A212F0L1 A0A2W1BIF8 A0A2J7PDR9 A0A2J7PDR8 A0A1J1I7B3 A0A1J1I3C5 A0A2J7PDS3 A0A067QQ01 A0A0C9QRM8 A0A0K8V5U5 A0A0L7KU90 A0A151X6H7 A0A0C9R937 A0A1B0CL03 A0A0M3QYH8 A0A0A1XI03 A0A158NUK2 A0A151J329 A0A195BFB3 A0A195FK32 E9IRV8 A0A0P8XYD2 A0A195C7T5 B3M2A1 B4JHE7 A0A034WPZ3 A0A0K8UY96 A0A1W4VCY5 A0A0R1E6X6 A0A0R1E6H2 F4WRE1 B4PM80 A0A1W4VD75 B4GDZ3 I5AP46 K7IMN5 A0A1A9ZM88 B0WKE7 A0A232ES00 A0A1A9YBN3 A0A0Q9WXW4 A0A1A9VV64 B4KC90 A0A0A1XKH1 A0A1B0AVD2 Q297H6 B4M5J0 A0A3B0KBY5 B4QWC0 A0A1L8DL63 B3P3I0 A0A1L8DLE7 B4IBM4 W8B222 A0A1I8N2F5 A0A026W436 A0A0J7KR71 A0A3L8DUK9 Q9VEN5 A0A154PBA4 Q7KSF5 B4NJZ3 A0A1I8NMX4 A9UNI1 A0A1I8NMX3 A0A1Y1LAL4 A0A0L7QSP2 W5JHV8 A0A182J6L3 A0A2A3EEH5 A0A2M4AE20 A0A2M4AEH8 A0A2M4AEG7 A0A2M4BBY6 A0A087ZSQ0 A0A2M4BBW3 A0A1B6E609 A0A1B6DL59 A0A182YG19 A0A1Y1LAN1 A0A1Y1LEC9 A0A1Y1LAL5 E0VSY6 A0A182KCB4 Q5TT67 A0A084VTM4 A0A182RFK7 A0A182NML2 A0A182WVL6 A0A1Y1LCT2 A0A182I995 A0A182Q9P9 A0A182FPQ9
A0A3S2NUV0 A0A212F0L1 A0A2W1BIF8 A0A2J7PDR9 A0A2J7PDR8 A0A1J1I7B3 A0A1J1I3C5 A0A2J7PDS3 A0A067QQ01 A0A0C9QRM8 A0A0K8V5U5 A0A0L7KU90 A0A151X6H7 A0A0C9R937 A0A1B0CL03 A0A0M3QYH8 A0A0A1XI03 A0A158NUK2 A0A151J329 A0A195BFB3 A0A195FK32 E9IRV8 A0A0P8XYD2 A0A195C7T5 B3M2A1 B4JHE7 A0A034WPZ3 A0A0K8UY96 A0A1W4VCY5 A0A0R1E6X6 A0A0R1E6H2 F4WRE1 B4PM80 A0A1W4VD75 B4GDZ3 I5AP46 K7IMN5 A0A1A9ZM88 B0WKE7 A0A232ES00 A0A1A9YBN3 A0A0Q9WXW4 A0A1A9VV64 B4KC90 A0A0A1XKH1 A0A1B0AVD2 Q297H6 B4M5J0 A0A3B0KBY5 B4QWC0 A0A1L8DL63 B3P3I0 A0A1L8DLE7 B4IBM4 W8B222 A0A1I8N2F5 A0A026W436 A0A0J7KR71 A0A3L8DUK9 Q9VEN5 A0A154PBA4 Q7KSF5 B4NJZ3 A0A1I8NMX4 A9UNI1 A0A1I8NMX3 A0A1Y1LAL4 A0A0L7QSP2 W5JHV8 A0A182J6L3 A0A2A3EEH5 A0A2M4AE20 A0A2M4AEH8 A0A2M4AEG7 A0A2M4BBY6 A0A087ZSQ0 A0A2M4BBW3 A0A1B6E609 A0A1B6DL59 A0A182YG19 A0A1Y1LAN1 A0A1Y1LEC9 A0A1Y1LAL5 E0VSY6 A0A182KCB4 Q5TT67 A0A084VTM4 A0A182RFK7 A0A182NML2 A0A182WVL6 A0A1Y1LCT2 A0A182I995 A0A182Q9P9 A0A182FPQ9
Pubmed
29482325
26354079
22118469
28756777
24845553
26227816
+ More
25830018 21347285 21282665 17994087 18057021 25348373 17550304 21719571 15632085 23185243 20075255 28648823 24495485 25315136 24508170 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 20920257 23761445 25244985 20566863 12364791 14747013 17210077 24438588
25830018 21347285 21282665 17994087 18057021 25348373 17550304 21719571 15632085 23185243 20075255 28648823 24495485 25315136 24508170 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 20920257 23761445 25244985 20566863 12364791 14747013 17210077 24438588
EMBL
MG860914
AVO00712.1
NWSH01001847
PCG69932.1
PCG69934.1
KU355788
+ More
APW79907.1 KQ459586 KPI98024.1 KQ460883 KPJ11255.1 RSAL01000068 RVE49292.1 AGBW02011073 OWR47270.1 KZ150103 PZC73474.1 NEVH01026157 PNF14484.1 PNF14485.1 CVRI01000040 CRK94838.1 CRK94839.1 PNF14486.1 KK853073 KDR11784.1 GBYB01003287 JAG73054.1 GDHF01033460 GDHF01018022 JAI18854.1 JAI34292.1 JTDY01005613 KOB66832.1 KQ982481 KYQ55993.1 GBYB01003286 JAG73053.1 AJWK01016819 AJWK01016820 AJWK01016821 AJWK01016822 CP012526 ALC47703.1 GBXI01003705 JAD10587.1 ADTU01026586 KQ980368 KYN16336.1 KQ976504 KYM82860.1 KQ981522 KYN40622.1 GL765283 EFZ16728.1 CH902617 KPU79645.1 KPU79647.1 KQ978143 KYM96710.1 EDV42292.1 CH916369 EDV93854.1 GAKP01003129 JAC55823.1 GDHF01020667 GDHF01019919 GDHF01018093 GDHF01006525 JAI31647.1 JAI32395.1 JAI34221.1 JAI45789.1 CM000160 KRK03454.1 KRK03455.1 KRK03458.1 KRK03459.1 KRK03457.1 GL888285 EGI63212.1 EDW96946.1 CH479182 EDW33828.1 CM000070 EIM52731.1 KRT00579.1 KRT00581.1 DS231971 EDS29766.1 NNAY01002537 OXU21112.1 CH933806 KRG00580.1 KRG00581.1 EDW13699.2 GBXI01002781 JAD11511.1 JXJN01004197 JXJN01004198 EAL28229.3 CH940652 EDW58916.2 OUUW01000007 SPP83206.1 CM000364 EDX12611.1 GFDF01006888 JAV07196.1 CH954181 EDV48760.1 GFDF01006889 JAV07195.1 CH480827 EDW44782.1 GAMC01011360 JAB95195.1 KK107496 EZA49809.1 LBMM01004124 KMQ92791.1 QOIP01000004 RLU24084.1 AE014297 AAF55386.1 AAO41571.1 KQ434868 KZC09189.1 DQ372684 BT099945 AAN13732.2 ABD14408.1 ACX37659.1 CH964272 EDW83995.1 BT031345 ABY21758.1 GEZM01061195 GEZM01061188 JAV70689.1 KQ414756 KOC61650.1 ADMH02001205 ETN63706.1 AXCP01007962 KZ288266 PBC30155.1 GGFK01005714 MBW39035.1 GGFK01005707 MBW39028.1 GGFK01005687 MBW39008.1 GGFJ01001396 MBW50537.1 GGFJ01001395 MBW50536.1 GEDC01008706 GEDC01003964 JAS28592.1 JAS33334.1 GEDC01029437 GEDC01023143 GEDC01010893 JAS07861.1 JAS14155.1 JAS26405.1 GEZM01061192 JAV70693.1 GEZM01061187 JAV70700.1 GEZM01061193 JAV70692.1 DS235758 EEB16492.1 AAAB01008888 EAL40607.4 ATLV01016361 KE525079 KFB41318.1 GEZM01061194 GEZM01061191 JAV70691.1 APCN01003236 APCN01003237 AXCN02001597
APW79907.1 KQ459586 KPI98024.1 KQ460883 KPJ11255.1 RSAL01000068 RVE49292.1 AGBW02011073 OWR47270.1 KZ150103 PZC73474.1 NEVH01026157 PNF14484.1 PNF14485.1 CVRI01000040 CRK94838.1 CRK94839.1 PNF14486.1 KK853073 KDR11784.1 GBYB01003287 JAG73054.1 GDHF01033460 GDHF01018022 JAI18854.1 JAI34292.1 JTDY01005613 KOB66832.1 KQ982481 KYQ55993.1 GBYB01003286 JAG73053.1 AJWK01016819 AJWK01016820 AJWK01016821 AJWK01016822 CP012526 ALC47703.1 GBXI01003705 JAD10587.1 ADTU01026586 KQ980368 KYN16336.1 KQ976504 KYM82860.1 KQ981522 KYN40622.1 GL765283 EFZ16728.1 CH902617 KPU79645.1 KPU79647.1 KQ978143 KYM96710.1 EDV42292.1 CH916369 EDV93854.1 GAKP01003129 JAC55823.1 GDHF01020667 GDHF01019919 GDHF01018093 GDHF01006525 JAI31647.1 JAI32395.1 JAI34221.1 JAI45789.1 CM000160 KRK03454.1 KRK03455.1 KRK03458.1 KRK03459.1 KRK03457.1 GL888285 EGI63212.1 EDW96946.1 CH479182 EDW33828.1 CM000070 EIM52731.1 KRT00579.1 KRT00581.1 DS231971 EDS29766.1 NNAY01002537 OXU21112.1 CH933806 KRG00580.1 KRG00581.1 EDW13699.2 GBXI01002781 JAD11511.1 JXJN01004197 JXJN01004198 EAL28229.3 CH940652 EDW58916.2 OUUW01000007 SPP83206.1 CM000364 EDX12611.1 GFDF01006888 JAV07196.1 CH954181 EDV48760.1 GFDF01006889 JAV07195.1 CH480827 EDW44782.1 GAMC01011360 JAB95195.1 KK107496 EZA49809.1 LBMM01004124 KMQ92791.1 QOIP01000004 RLU24084.1 AE014297 AAF55386.1 AAO41571.1 KQ434868 KZC09189.1 DQ372684 BT099945 AAN13732.2 ABD14408.1 ACX37659.1 CH964272 EDW83995.1 BT031345 ABY21758.1 GEZM01061195 GEZM01061188 JAV70689.1 KQ414756 KOC61650.1 ADMH02001205 ETN63706.1 AXCP01007962 KZ288266 PBC30155.1 GGFK01005714 MBW39035.1 GGFK01005707 MBW39028.1 GGFK01005687 MBW39008.1 GGFJ01001396 MBW50537.1 GGFJ01001395 MBW50536.1 GEDC01008706 GEDC01003964 JAS28592.1 JAS33334.1 GEDC01029437 GEDC01023143 GEDC01010893 JAS07861.1 JAS14155.1 JAS26405.1 GEZM01061192 JAV70693.1 GEZM01061187 JAV70700.1 GEZM01061193 JAV70692.1 DS235758 EEB16492.1 AAAB01008888 EAL40607.4 ATLV01016361 KE525079 KFB41318.1 GEZM01061194 GEZM01061191 JAV70691.1 APCN01003236 APCN01003237 AXCN02001597
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
UP000235965
+ More
UP000183832 UP000027135 UP000037510 UP000075809 UP000092461 UP000092553 UP000005205 UP000078492 UP000078540 UP000078541 UP000007801 UP000078542 UP000001070 UP000192221 UP000002282 UP000007755 UP000008744 UP000001819 UP000002358 UP000092445 UP000002320 UP000215335 UP000092443 UP000009192 UP000078200 UP000092460 UP000008792 UP000268350 UP000000304 UP000008711 UP000001292 UP000095301 UP000053097 UP000036403 UP000279307 UP000000803 UP000076502 UP000007798 UP000095300 UP000053825 UP000000673 UP000075880 UP000242457 UP000005203 UP000076408 UP000009046 UP000075881 UP000007062 UP000030765 UP000075900 UP000075884 UP000076407 UP000075840 UP000075886 UP000069272
UP000183832 UP000027135 UP000037510 UP000075809 UP000092461 UP000092553 UP000005205 UP000078492 UP000078540 UP000078541 UP000007801 UP000078542 UP000001070 UP000192221 UP000002282 UP000007755 UP000008744 UP000001819 UP000002358 UP000092445 UP000002320 UP000215335 UP000092443 UP000009192 UP000078200 UP000092460 UP000008792 UP000268350 UP000000304 UP000008711 UP000001292 UP000095301 UP000053097 UP000036403 UP000279307 UP000000803 UP000076502 UP000007798 UP000095300 UP000053825 UP000000673 UP000075880 UP000242457 UP000005203 UP000076408 UP000009046 UP000075881 UP000007062 UP000030765 UP000075900 UP000075884 UP000076407 UP000075840 UP000075886 UP000069272
Interpro
Gene 3D
ProteinModelPortal
A0A2P1JMP2
A0A2A4JEU7
A0A2A4JEC7
A0A2H4G7J0
A0A194PZ49
A0A194R1X7
+ More
A0A3S2NUV0 A0A212F0L1 A0A2W1BIF8 A0A2J7PDR9 A0A2J7PDR8 A0A1J1I7B3 A0A1J1I3C5 A0A2J7PDS3 A0A067QQ01 A0A0C9QRM8 A0A0K8V5U5 A0A0L7KU90 A0A151X6H7 A0A0C9R937 A0A1B0CL03 A0A0M3QYH8 A0A0A1XI03 A0A158NUK2 A0A151J329 A0A195BFB3 A0A195FK32 E9IRV8 A0A0P8XYD2 A0A195C7T5 B3M2A1 B4JHE7 A0A034WPZ3 A0A0K8UY96 A0A1W4VCY5 A0A0R1E6X6 A0A0R1E6H2 F4WRE1 B4PM80 A0A1W4VD75 B4GDZ3 I5AP46 K7IMN5 A0A1A9ZM88 B0WKE7 A0A232ES00 A0A1A9YBN3 A0A0Q9WXW4 A0A1A9VV64 B4KC90 A0A0A1XKH1 A0A1B0AVD2 Q297H6 B4M5J0 A0A3B0KBY5 B4QWC0 A0A1L8DL63 B3P3I0 A0A1L8DLE7 B4IBM4 W8B222 A0A1I8N2F5 A0A026W436 A0A0J7KR71 A0A3L8DUK9 Q9VEN5 A0A154PBA4 Q7KSF5 B4NJZ3 A0A1I8NMX4 A9UNI1 A0A1I8NMX3 A0A1Y1LAL4 A0A0L7QSP2 W5JHV8 A0A182J6L3 A0A2A3EEH5 A0A2M4AE20 A0A2M4AEH8 A0A2M4AEG7 A0A2M4BBY6 A0A087ZSQ0 A0A2M4BBW3 A0A1B6E609 A0A1B6DL59 A0A182YG19 A0A1Y1LAN1 A0A1Y1LEC9 A0A1Y1LAL5 E0VSY6 A0A182KCB4 Q5TT67 A0A084VTM4 A0A182RFK7 A0A182NML2 A0A182WVL6 A0A1Y1LCT2 A0A182I995 A0A182Q9P9 A0A182FPQ9
A0A3S2NUV0 A0A212F0L1 A0A2W1BIF8 A0A2J7PDR9 A0A2J7PDR8 A0A1J1I7B3 A0A1J1I3C5 A0A2J7PDS3 A0A067QQ01 A0A0C9QRM8 A0A0K8V5U5 A0A0L7KU90 A0A151X6H7 A0A0C9R937 A0A1B0CL03 A0A0M3QYH8 A0A0A1XI03 A0A158NUK2 A0A151J329 A0A195BFB3 A0A195FK32 E9IRV8 A0A0P8XYD2 A0A195C7T5 B3M2A1 B4JHE7 A0A034WPZ3 A0A0K8UY96 A0A1W4VCY5 A0A0R1E6X6 A0A0R1E6H2 F4WRE1 B4PM80 A0A1W4VD75 B4GDZ3 I5AP46 K7IMN5 A0A1A9ZM88 B0WKE7 A0A232ES00 A0A1A9YBN3 A0A0Q9WXW4 A0A1A9VV64 B4KC90 A0A0A1XKH1 A0A1B0AVD2 Q297H6 B4M5J0 A0A3B0KBY5 B4QWC0 A0A1L8DL63 B3P3I0 A0A1L8DLE7 B4IBM4 W8B222 A0A1I8N2F5 A0A026W436 A0A0J7KR71 A0A3L8DUK9 Q9VEN5 A0A154PBA4 Q7KSF5 B4NJZ3 A0A1I8NMX4 A9UNI1 A0A1I8NMX3 A0A1Y1LAL4 A0A0L7QSP2 W5JHV8 A0A182J6L3 A0A2A3EEH5 A0A2M4AE20 A0A2M4AEH8 A0A2M4AEG7 A0A2M4BBY6 A0A087ZSQ0 A0A2M4BBW3 A0A1B6E609 A0A1B6DL59 A0A182YG19 A0A1Y1LAN1 A0A1Y1LEC9 A0A1Y1LAL5 E0VSY6 A0A182KCB4 Q5TT67 A0A084VTM4 A0A182RFK7 A0A182NML2 A0A182WVL6 A0A1Y1LCT2 A0A182I995 A0A182Q9P9 A0A182FPQ9
PDB
4HXI
E-value=4.60784e-38,
Score=396
Ontologies
GO
Topology
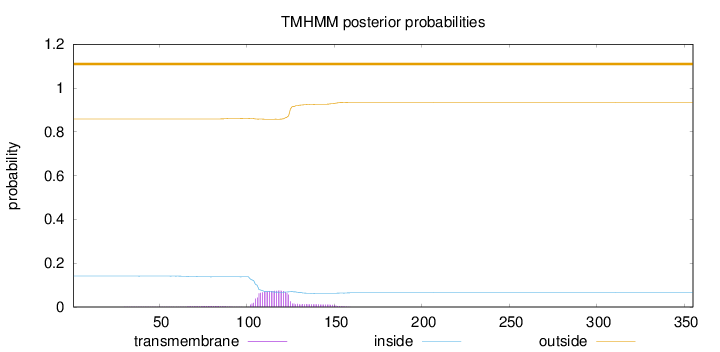
Length:
355
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.82956
Exp number, first 60 AAs:
0.00996
Total prob of N-in:
0.14142
outside
1 - 355
Population Genetic Test Statistics
Pi
182.72467
Theta
166.475991
Tajima's D
0.439288
CLR
0.519691
CSRT
0.497125143742813
Interpretation
Uncertain
