Gene
KWMTBOMO13306
Pre Gene Modal
BGIBMGA004700
Annotation
PREDICTED:_dual_specificity_protein_phosphatase_7_[Bombyx_mori]
Full name
Dual specificity protein phosphatase
+ More
Dual specificity protein phosphatase Mpk3
Dual specificity protein phosphatase Mpk3
Alternative Name
Drosophila MKP3
Mitogen-activated protein kinase phosphatase 3
Mitogen-activated protein kinase phosphatase 3
Location in the cell
Extracellular Reliability : 1.229 Nuclear Reliability : 1.696 PlasmaMembrane Reliability : 1.045
Sequence
CDS
ATGCTTAGGCGGTTAGCTGCTGGGAAGATTGAGTTGTCATCAACAGTGCAGTGCAAAGAGTTGAAGGCGAGGATAAACCACTGTCGAACCCGTGGTATATTCGTGTTGTACGGTGATGGACCTCCCAGAGACCCTGACTCTGTCCACGGTATACTCCTCAAGAGGCTGAAGCAAGACAGCGTGCAAGTCGTTTGTCTTGAAGGTGACTTCGATGACTTCCGTCGCGCATACCCCGAATGGTGCAGCGAAGCTGGCGCCCAACATGTACCCCACCTGCCGCTAATGGGATTACGGTCGCTGCGTATTTCGGGTTCGGGTTGCGAGGAGACCCTGTCCTCTGGCTCGTCGTCGGAGTGCGAGGAGGTGCACAGCCACGTCCAGCAGGACTTCCCTATCGAGATCCTGCCGAATCTGTACCTCGGTAACTCCACCAACAGCGAGGATTGCGAAGCTCTCGCCAGGCATAATATTAAGGTGATTATTCTATTCTGTGTGCTTAGTGCGAGTTTCTTAACGTTTTCGATAACGTAA
Protein
MLRRLAAGKIELSSTVQCKELKARINHCRTRGIFVLYGDGPPRDPDSVHGILLKRLKQDSVQVVCLEGDFDDFRRAYPEWCSEAGAQHVPHLPLMGLRSLRISGSGCEETLSSGSSSECEEVHSHVQQDFPIEILPNLYLGNSTNSEDCEALARHNIKVIILFCVLSASFLTFSIT
Summary
Description
Negatively regulates the activity of members of the MAP kinase family in response to changes in the cellular environment. Has a specificity for the ERK family. Acts as negative regulator in a variety of developmental processes including cell differentiation and proliferation controlled by the Ras/ERK pathway. Suppresses the photoreceptor cell differentiation and wing vein formation. Required for proper oogenesis and early embryogenesis. Functions autonomously in a subset of photoreceptor progenitor cells in eye imaginal disks. Appears also to be required in surrounding non-neuronal cells for ommatidial patterning and photoreceptor differentiation. Plays a role in the maintenance of epithelial integrity during tracheal development.
Catalytic Activity
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
Subunit
Interacts (via N-terminal region) with phosphorylated rl.
Similarity
Belongs to the protein-tyrosine phosphatase family. Non-receptor class dual specificity subfamily.
Keywords
Alternative splicing
Complete proteome
Cytoplasm
Hydrolase
Protein phosphatase
Reference proteome
Feature
chain Dual specificity protein phosphatase Mpk3
splice variant In isoform A.
splice variant In isoform A.
Uniprot
A0A0L7LQ89
S4P960
A0A2W1BE91
A0A212F0I3
A0A3S2LAE9
A0A067R934
+ More
A0A1B6L609 A0A1B6I4H5 A0A1B6F2Q1 A0A023F2P3 A0A1B6BWD5 E0VAX6 T1I5F2 A0A1B0CKU5 A0A1L8E2X3 D6WZI5 A0A026W176 V5GZC3 A0A1Y1LV60 A0A1Y1LWH0 E2A378 K7IM78 E2C241 A0A0A9YX42 A0A0A9YYM7 F4WRD4 A0A151J325 A0A151I2K5 A0A158NUJ4 A0A195C9C7 A0A087ZSQ5 A0A2A3EGN0 A0A2A3EFU8 A0A151WKA1 A0A1S4G7S1 A0A195FKV8 A0A0L7QSN8 E9IRV1 A0A2M4CM13 A0A1Q3FUD0 Q7PV94 A0A0M4EMY2 T1PAJ6 A0A154PBF2 A0A2M4APE5 A0A2S2Q8S1 A0A0M9A1T5 A0A0K8WK77 A0A2M4AAE0 A0A2H8TPQ7 A0A084WB53 A0A034VQ01 A0A1I8N269 U5EXJ8 A0A0A1WL42 M9PFW0 Q9VVW5 A0A2C9GQA1 B3NDX2 B4IFZ3 B4KZ97 M9PG13 W8ADW6 B4LD28 B4PH00 B4QQC5 A0A1B0FBR6 A0A1J1HUD1 B4IY47 A0A0L0C8K9 J9K9N6 A0A1S3IT30 B3M7P3 A0A336M0Y7 A0A1W4UP65 A0A1I8QBA9 B4H6S8 Q29DI8 A0A087UFB5 A0A3B0JM89 B4N4R1 A0A3B0JS64 A0A0P4W116 T1GQ64 A0A2L2YBF8 A0A2T7NUM3 T1JDA7 A0A0B6ZWL2 A0A2C9JCT1 R7TQ03
A0A1B6L609 A0A1B6I4H5 A0A1B6F2Q1 A0A023F2P3 A0A1B6BWD5 E0VAX6 T1I5F2 A0A1B0CKU5 A0A1L8E2X3 D6WZI5 A0A026W176 V5GZC3 A0A1Y1LV60 A0A1Y1LWH0 E2A378 K7IM78 E2C241 A0A0A9YX42 A0A0A9YYM7 F4WRD4 A0A151J325 A0A151I2K5 A0A158NUJ4 A0A195C9C7 A0A087ZSQ5 A0A2A3EGN0 A0A2A3EFU8 A0A151WKA1 A0A1S4G7S1 A0A195FKV8 A0A0L7QSN8 E9IRV1 A0A2M4CM13 A0A1Q3FUD0 Q7PV94 A0A0M4EMY2 T1PAJ6 A0A154PBF2 A0A2M4APE5 A0A2S2Q8S1 A0A0M9A1T5 A0A0K8WK77 A0A2M4AAE0 A0A2H8TPQ7 A0A084WB53 A0A034VQ01 A0A1I8N269 U5EXJ8 A0A0A1WL42 M9PFW0 Q9VVW5 A0A2C9GQA1 B3NDX2 B4IFZ3 B4KZ97 M9PG13 W8ADW6 B4LD28 B4PH00 B4QQC5 A0A1B0FBR6 A0A1J1HUD1 B4IY47 A0A0L0C8K9 J9K9N6 A0A1S3IT30 B3M7P3 A0A336M0Y7 A0A1W4UP65 A0A1I8QBA9 B4H6S8 Q29DI8 A0A087UFB5 A0A3B0JM89 B4N4R1 A0A3B0JS64 A0A0P4W116 T1GQ64 A0A2L2YBF8 A0A2T7NUM3 T1JDA7 A0A0B6ZWL2 A0A2C9JCT1 R7TQ03
EC Number
3.1.3.16
3.1.3.48
3.1.3.48
Pubmed
26227816
23622113
28756777
22118469
24845553
25474469
+ More
20566863 18362917 19820115 24508170 30249741 28004739 20798317 20075255 25401762 26823975 21719571 21347285 17510324 21282665 12364791 24438588 25348373 25315136 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11742539 12537569 15033693 12810595 14701731 15704110 16831830 17994087 24495485 17550304 26108605 15632085 26561354 15562597 23254933
20566863 18362917 19820115 24508170 30249741 28004739 20798317 20075255 25401762 26823975 21719571 21347285 17510324 21282665 12364791 24438588 25348373 25315136 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11742539 12537569 15033693 12810595 14701731 15704110 16831830 17994087 24495485 17550304 26108605 15632085 26561354 15562597 23254933
EMBL
JTDY01000398
KOB77366.1
GAIX01005661
JAA86899.1
KZ150103
PZC73472.1
+ More
AGBW02011073 OWR47268.1 RSAL01000068 RVE49293.1 KK852616 KDR20183.1 GEBQ01020828 JAT19149.1 GECU01025870 JAS81836.1 GECZ01025304 JAS44465.1 GBBI01003316 JAC15396.1 GEDC01031696 JAS05602.1 DS235019 EEB10532.1 ACPB03011499 ACPB03011500 AJWK01016641 AJWK01016642 AJWK01016643 AJWK01016644 AJWK01016645 AJWK01016646 AJWK01016647 AJWK01016648 AJWK01016649 GFDF01000991 JAV13093.1 KQ971372 EFA10433.2 KK107496 QOIP01000004 EZA49817.1 RLU24209.1 GALX01002623 JAB65843.1 GEZM01046135 GEZM01046134 JAV77492.1 GEZM01046136 JAV77491.1 GL436311 EFN72106.1 GL452067 EFN77992.1 GBHO01007407 GBHO01007403 GDHC01012020 JAG36197.1 JAG36201.1 JAQ06609.1 GBHO01007408 GBHO01007406 GBRD01014408 GDHC01017980 GDHC01015251 JAG36196.1 JAG36198.1 JAG51418.1 JAQ00649.1 JAQ03378.1 GL888285 EGI63205.1 KQ980368 KYN16326.1 KQ976533 KYM81495.1 ADTU01026584 KQ978143 KYM96718.1 KZ288266 PBC30161.1 PBC30162.1 KQ983014 KYQ48240.1 KQ981522 KYN40629.1 KQ414756 KOC61645.1 GL765283 EFZ16686.1 GGFL01001750 MBW65928.1 GFDL01003959 JAV31086.1 AAAB01008986 EAA00236.5 CP012525 ALC45147.1 KA645757 AFP60386.1 KQ434868 KZC09182.1 GGFK01009157 MBW42478.1 GGMS01004926 MBY74129.1 KQ435760 KOX75570.1 GDHF01028712 GDHF01017961 GDHF01016312 GDHF01000850 JAI23602.1 JAI34353.1 JAI36002.1 JAI51464.1 GGFK01004442 MBW37763.1 GFXV01003423 MBW15228.1 ATLV01022313 ATLV01022314 KE525331 KFB47447.1 GAKP01013576 JAC45376.1 GANO01000051 JAB59820.1 GBXI01015057 JAC99234.1 AE014296 AGB94725.1 AY043264 AY060472 BT003536 AAF49192.2 AAF49193.2 AAO39540.1 APCN01003283 APCN01003284 CH954178 EDV52326.2 CH480834 EDW46580.1 CH933809 EDW18923.2 AGB94724.1 GAMC01020218 GAMC01020215 JAB86340.1 CH940647 EDW69909.2 CM000159 EDW95370.2 CM000363 EDX11031.1 CCAG010004107 CVRI01000021 CRK91679.1 CH916366 EDV97590.1 JRES01000760 KNC28567.1 ABLF02027754 ABLF02027755 CH902618 EDV40971.2 UFQT01000391 SSX23892.1 CH479215 EDW33516.1 CH379070 EAL30426.3 KK119571 KFM76054.1 OUUW01000002 SPP76550.1 CH964095 EDW79135.2 SPP76549.1 GDRN01081109 JAI62078.1 CAQQ02189220 IAAA01010301 LAA05496.1 PZQS01000009 PVD24854.1 JH432102 HACG01026074 HACG01026075 CEK72939.1 CEK72940.1 AMQN01011685 KB309028 ELT95734.1
AGBW02011073 OWR47268.1 RSAL01000068 RVE49293.1 KK852616 KDR20183.1 GEBQ01020828 JAT19149.1 GECU01025870 JAS81836.1 GECZ01025304 JAS44465.1 GBBI01003316 JAC15396.1 GEDC01031696 JAS05602.1 DS235019 EEB10532.1 ACPB03011499 ACPB03011500 AJWK01016641 AJWK01016642 AJWK01016643 AJWK01016644 AJWK01016645 AJWK01016646 AJWK01016647 AJWK01016648 AJWK01016649 GFDF01000991 JAV13093.1 KQ971372 EFA10433.2 KK107496 QOIP01000004 EZA49817.1 RLU24209.1 GALX01002623 JAB65843.1 GEZM01046135 GEZM01046134 JAV77492.1 GEZM01046136 JAV77491.1 GL436311 EFN72106.1 GL452067 EFN77992.1 GBHO01007407 GBHO01007403 GDHC01012020 JAG36197.1 JAG36201.1 JAQ06609.1 GBHO01007408 GBHO01007406 GBRD01014408 GDHC01017980 GDHC01015251 JAG36196.1 JAG36198.1 JAG51418.1 JAQ00649.1 JAQ03378.1 GL888285 EGI63205.1 KQ980368 KYN16326.1 KQ976533 KYM81495.1 ADTU01026584 KQ978143 KYM96718.1 KZ288266 PBC30161.1 PBC30162.1 KQ983014 KYQ48240.1 KQ981522 KYN40629.1 KQ414756 KOC61645.1 GL765283 EFZ16686.1 GGFL01001750 MBW65928.1 GFDL01003959 JAV31086.1 AAAB01008986 EAA00236.5 CP012525 ALC45147.1 KA645757 AFP60386.1 KQ434868 KZC09182.1 GGFK01009157 MBW42478.1 GGMS01004926 MBY74129.1 KQ435760 KOX75570.1 GDHF01028712 GDHF01017961 GDHF01016312 GDHF01000850 JAI23602.1 JAI34353.1 JAI36002.1 JAI51464.1 GGFK01004442 MBW37763.1 GFXV01003423 MBW15228.1 ATLV01022313 ATLV01022314 KE525331 KFB47447.1 GAKP01013576 JAC45376.1 GANO01000051 JAB59820.1 GBXI01015057 JAC99234.1 AE014296 AGB94725.1 AY043264 AY060472 BT003536 AAF49192.2 AAF49193.2 AAO39540.1 APCN01003283 APCN01003284 CH954178 EDV52326.2 CH480834 EDW46580.1 CH933809 EDW18923.2 AGB94724.1 GAMC01020218 GAMC01020215 JAB86340.1 CH940647 EDW69909.2 CM000159 EDW95370.2 CM000363 EDX11031.1 CCAG010004107 CVRI01000021 CRK91679.1 CH916366 EDV97590.1 JRES01000760 KNC28567.1 ABLF02027754 ABLF02027755 CH902618 EDV40971.2 UFQT01000391 SSX23892.1 CH479215 EDW33516.1 CH379070 EAL30426.3 KK119571 KFM76054.1 OUUW01000002 SPP76550.1 CH964095 EDW79135.2 SPP76549.1 GDRN01081109 JAI62078.1 CAQQ02189220 IAAA01010301 LAA05496.1 PZQS01000009 PVD24854.1 JH432102 HACG01026074 HACG01026075 CEK72939.1 CEK72940.1 AMQN01011685 KB309028 ELT95734.1
Proteomes
UP000037510
UP000007151
UP000283053
UP000027135
UP000009046
UP000015103
+ More
UP000092461 UP000007266 UP000053097 UP000279307 UP000000311 UP000002358 UP000008237 UP000007755 UP000078492 UP000078540 UP000005205 UP000078542 UP000005203 UP000242457 UP000075809 UP000078541 UP000053825 UP000007062 UP000092553 UP000076502 UP000053105 UP000030765 UP000095301 UP000000803 UP000075840 UP000008711 UP000001292 UP000009192 UP000008792 UP000002282 UP000000304 UP000092444 UP000183832 UP000001070 UP000037069 UP000007819 UP000085678 UP000007801 UP000192221 UP000095300 UP000008744 UP000001819 UP000054359 UP000268350 UP000007798 UP000015102 UP000245119 UP000076420 UP000014760
UP000092461 UP000007266 UP000053097 UP000279307 UP000000311 UP000002358 UP000008237 UP000007755 UP000078492 UP000078540 UP000005205 UP000078542 UP000005203 UP000242457 UP000075809 UP000078541 UP000053825 UP000007062 UP000092553 UP000076502 UP000053105 UP000030765 UP000095301 UP000000803 UP000075840 UP000008711 UP000001292 UP000009192 UP000008792 UP000002282 UP000000304 UP000092444 UP000183832 UP000001070 UP000037069 UP000007819 UP000085678 UP000007801 UP000192221 UP000095300 UP000008744 UP000001819 UP000054359 UP000268350 UP000007798 UP000015102 UP000245119 UP000076420 UP000014760
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A0L7LQ89
S4P960
A0A2W1BE91
A0A212F0I3
A0A3S2LAE9
A0A067R934
+ More
A0A1B6L609 A0A1B6I4H5 A0A1B6F2Q1 A0A023F2P3 A0A1B6BWD5 E0VAX6 T1I5F2 A0A1B0CKU5 A0A1L8E2X3 D6WZI5 A0A026W176 V5GZC3 A0A1Y1LV60 A0A1Y1LWH0 E2A378 K7IM78 E2C241 A0A0A9YX42 A0A0A9YYM7 F4WRD4 A0A151J325 A0A151I2K5 A0A158NUJ4 A0A195C9C7 A0A087ZSQ5 A0A2A3EGN0 A0A2A3EFU8 A0A151WKA1 A0A1S4G7S1 A0A195FKV8 A0A0L7QSN8 E9IRV1 A0A2M4CM13 A0A1Q3FUD0 Q7PV94 A0A0M4EMY2 T1PAJ6 A0A154PBF2 A0A2M4APE5 A0A2S2Q8S1 A0A0M9A1T5 A0A0K8WK77 A0A2M4AAE0 A0A2H8TPQ7 A0A084WB53 A0A034VQ01 A0A1I8N269 U5EXJ8 A0A0A1WL42 M9PFW0 Q9VVW5 A0A2C9GQA1 B3NDX2 B4IFZ3 B4KZ97 M9PG13 W8ADW6 B4LD28 B4PH00 B4QQC5 A0A1B0FBR6 A0A1J1HUD1 B4IY47 A0A0L0C8K9 J9K9N6 A0A1S3IT30 B3M7P3 A0A336M0Y7 A0A1W4UP65 A0A1I8QBA9 B4H6S8 Q29DI8 A0A087UFB5 A0A3B0JM89 B4N4R1 A0A3B0JS64 A0A0P4W116 T1GQ64 A0A2L2YBF8 A0A2T7NUM3 T1JDA7 A0A0B6ZWL2 A0A2C9JCT1 R7TQ03
A0A1B6L609 A0A1B6I4H5 A0A1B6F2Q1 A0A023F2P3 A0A1B6BWD5 E0VAX6 T1I5F2 A0A1B0CKU5 A0A1L8E2X3 D6WZI5 A0A026W176 V5GZC3 A0A1Y1LV60 A0A1Y1LWH0 E2A378 K7IM78 E2C241 A0A0A9YX42 A0A0A9YYM7 F4WRD4 A0A151J325 A0A151I2K5 A0A158NUJ4 A0A195C9C7 A0A087ZSQ5 A0A2A3EGN0 A0A2A3EFU8 A0A151WKA1 A0A1S4G7S1 A0A195FKV8 A0A0L7QSN8 E9IRV1 A0A2M4CM13 A0A1Q3FUD0 Q7PV94 A0A0M4EMY2 T1PAJ6 A0A154PBF2 A0A2M4APE5 A0A2S2Q8S1 A0A0M9A1T5 A0A0K8WK77 A0A2M4AAE0 A0A2H8TPQ7 A0A084WB53 A0A034VQ01 A0A1I8N269 U5EXJ8 A0A0A1WL42 M9PFW0 Q9VVW5 A0A2C9GQA1 B3NDX2 B4IFZ3 B4KZ97 M9PG13 W8ADW6 B4LD28 B4PH00 B4QQC5 A0A1B0FBR6 A0A1J1HUD1 B4IY47 A0A0L0C8K9 J9K9N6 A0A1S3IT30 B3M7P3 A0A336M0Y7 A0A1W4UP65 A0A1I8QBA9 B4H6S8 Q29DI8 A0A087UFB5 A0A3B0JM89 B4N4R1 A0A3B0JS64 A0A0P4W116 T1GQ64 A0A2L2YBF8 A0A2T7NUM3 T1JDA7 A0A0B6ZWL2 A0A2C9JCT1 R7TQ03
PDB
2HXP
E-value=0.00164308,
Score=93
Ontologies
GO
Topology
Subcellular location
Cytoplasm
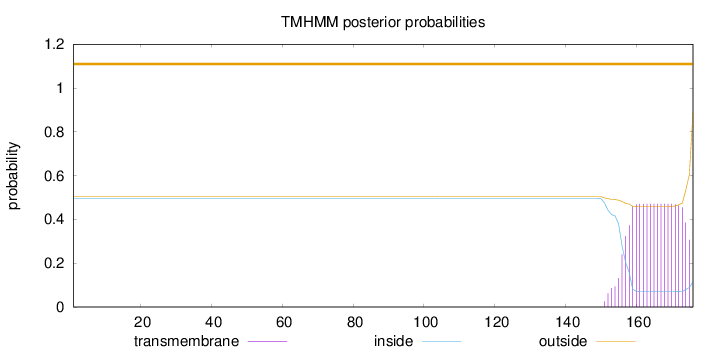
Length:
176
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.05561
Exp number, first 60 AAs:
0
Total prob of N-in:
0.49528
outside
1 - 176
Population Genetic Test Statistics
Pi
193.694506
Theta
179.099342
Tajima's D
0.504529
CLR
0.113905
CSRT
0.520173991300435
Interpretation
Uncertain
