Gene
KWMTBOMO13302
Pre Gene Modal
BGIBMGA004702
Annotation
PREDICTED:_transmembrane_protein_35_isoform_X2_[Papilio_xuthus]
Full name
NADH-cytochrome b5 reductase
Location in the cell
PlasmaMembrane Reliability : 1.908
Sequence
CDS
ATGATATCGAAAGAATTGCATAAGGATTTGAGAAAGGAGTATGTTAGATATGCAAAAGTATTTCCACTGACTGATACGTTGGACTTCAAATTACCATCGAAATGGTACAGGAGGACCGTGGGTGGTTTAGAAATATTATTTGGGTTAGCGTTGGCAGTTGCACCTAATCATAAGTTGAAGAACGCCGCGAACATCGGTCTGGTGTTGCTAATGATCCTGGCGGCGTACTCTCACGTGATGGTGGGCGACCCGTTCGACCGATGTGCGCCGGCTCTCGTATTCTTTTTCATGCTCAGTGGGAGGCTCGTCGTTTGGTATCAGACGAGTCGACGTGAACAGCTAGAGAAGTCCGCCACAGCCCAGAACGGCAACGGACTAAAGAGGGATTAA
Protein
MISKELHKDLRKEYVRYAKVFPLTDTLDFKLPSKWYRRTVGGLEILFGLALAVAPNHKLKNAANIGLVLLMILAAYSHVMVGDPFDRCAPALVFFFMLSGRLVVWYQTSRREQLEKSATAQNGNGLKRD
Summary
Catalytic Activity
2 [Fe(III)-cytochrome b5] + NADH = 2 [Fe(II)-cytochrome b5] + H(+) + NAD(+)
Cofactor
FAD
Similarity
Belongs to the flavoprotein pyridine nucleotide cytochrome reductase family.
Uniprot
A0A194PXE3
A0A212F0N4
A0A194R0N4
A0A2H1VWW1
A0A2W1BID4
H9J5B2
+ More
A0A023EH56 A0A182RSQ0 A0A1Q3FZK6 A0A182WJ68 A0A336LU73 Q7PT42 A0A182KRD5 A0A182Q2J9 A0A182ITK3 A0A182XYX9 T1EA69 Q16W86 A0A0L0C7H7 A0A034VRC8 A0A0C9R1P8 A0A0K8UA00 A0A182NHT8 B4N712 A0A1L8DA13 A0A1I8PR35 B3M7R9 B4PDR0 A0A0A1XHW1 A0A182UFU8 B3NES7 N6ULU9 A0A084WNC5 A0A1W4VV60 E9IK90 A0A182UZY9 A0A182KES5 A0A182PQD5 A0A182I3C5 B4H3G3 Q2LZP8 A0A182X3P4 A0A3B0JUD0 B4HVX2 B4QMH9 Q8T0T9 A0A139WI43 A0A1Y1K1H0 A0A1I8MBB6 A0A151JA11 A0A1B0FM45 A0A1A9Y9M3 A0A1A9VYK9 A0A1B0B9A0 A0A1A9Z3D5 W5JAN0 A0A182FF60 A0A0L7LTG0 T1I5P7 A0A067QQZ6 A0A023F9R2 E0W343 B4L974 R4G5E4 A0A0M3QWM3 A0A2A3E3D9 A0A0A9XZL1 A0A088A6Z7 V9ICU9 A0A195CSX8 A0A146KQV3 B4LBT9 A0A310SBE3 A0A154P7G6 A0A0M8ZNB0 E2BUW8 A0A195BX10 A0A158P3M8 A0A026WUE9 A0A0L7R561 E2AJ43 A0A195F192 B4J1X4 A0A1A9X2W2 A0A0K8TLH2 A0A1B6IW00 A0A1B6FDR1 A0A0T6BGG1 K7ITS8 A0A1B6E1Y9 D6WJE9 A0A1B0DPS7 A0A1B0CBS9 A0A182M9S9 A0A2H8TTE1 J9K3Z3 C4WTP6 A0A1D2MNR0 A0A2P2IAE9 A0A2R7WLI4
A0A023EH56 A0A182RSQ0 A0A1Q3FZK6 A0A182WJ68 A0A336LU73 Q7PT42 A0A182KRD5 A0A182Q2J9 A0A182ITK3 A0A182XYX9 T1EA69 Q16W86 A0A0L0C7H7 A0A034VRC8 A0A0C9R1P8 A0A0K8UA00 A0A182NHT8 B4N712 A0A1L8DA13 A0A1I8PR35 B3M7R9 B4PDR0 A0A0A1XHW1 A0A182UFU8 B3NES7 N6ULU9 A0A084WNC5 A0A1W4VV60 E9IK90 A0A182UZY9 A0A182KES5 A0A182PQD5 A0A182I3C5 B4H3G3 Q2LZP8 A0A182X3P4 A0A3B0JUD0 B4HVX2 B4QMH9 Q8T0T9 A0A139WI43 A0A1Y1K1H0 A0A1I8MBB6 A0A151JA11 A0A1B0FM45 A0A1A9Y9M3 A0A1A9VYK9 A0A1B0B9A0 A0A1A9Z3D5 W5JAN0 A0A182FF60 A0A0L7LTG0 T1I5P7 A0A067QQZ6 A0A023F9R2 E0W343 B4L974 R4G5E4 A0A0M3QWM3 A0A2A3E3D9 A0A0A9XZL1 A0A088A6Z7 V9ICU9 A0A195CSX8 A0A146KQV3 B4LBT9 A0A310SBE3 A0A154P7G6 A0A0M8ZNB0 E2BUW8 A0A195BX10 A0A158P3M8 A0A026WUE9 A0A0L7R561 E2AJ43 A0A195F192 B4J1X4 A0A1A9X2W2 A0A0K8TLH2 A0A1B6IW00 A0A1B6FDR1 A0A0T6BGG1 K7ITS8 A0A1B6E1Y9 D6WJE9 A0A1B0DPS7 A0A1B0CBS9 A0A182M9S9 A0A2H8TTE1 J9K3Z3 C4WTP6 A0A1D2MNR0 A0A2P2IAE9 A0A2R7WLI4
EC Number
1.6.2.2
Pubmed
26354079
22118469
28756777
19121390
24945155
12364791
+ More
14747013 17210077 20966253 25244985 17510324 26108605 25348373 17994087 17550304 25830018 23537049 24438588 21282665 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 28004739 25315136 20920257 23761445 26227816 24845553 25474469 20566863 25401762 26823975 20798317 21347285 24508170 30249741 26369729 20075255 27289101
14747013 17210077 20966253 25244985 17510324 26108605 25348373 17994087 17550304 25830018 23537049 24438588 21282665 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 28004739 25315136 20920257 23761445 26227816 24845553 25474469 20566863 25401762 26823975 20798317 21347285 24508170 30249741 26369729 20075255 27289101
EMBL
KQ459586
KPI98016.1
AGBW02011073
OWR47261.1
KQ460883
KPJ11262.1
+ More
ODYU01004946 SOQ45337.1 KZ150103 PZC73465.1 BABH01037020 GAPW01005393 JAC08205.1 GFDL01002057 JAV32988.1 UFQS01000127 UFQT01000127 SSX00161.1 SSX20541.1 AAAB01008807 EAA04565.5 AXCN02000377 GAMD01000697 JAB00894.1 CH477572 EAT38864.1 JRES01000810 KNC28201.1 GAKP01014844 JAC44108.1 GBYB01000761 JAG70528.1 GDHF01028906 GDHF01022416 GDHF01011113 JAI23408.1 JAI29898.1 JAI41201.1 CH964168 EDW80151.1 GFDF01010914 JAV03170.1 CH902618 EDV38792.1 CM000159 EDW92875.1 GBXI01004199 JAD10093.1 CH954178 EDV50200.1 APGK01027575 APGK01027576 APGK01027577 APGK01027578 APGK01027579 KB740664 KB631791 ENN79682.1 ERL86099.1 ATLV01024587 KE525352 KFB51719.1 GL763924 EFZ19021.1 APCN01000205 CH479206 EDW30903.1 CH379069 EAL29459.2 KRT08027.1 OUUW01000002 SPP76966.1 CH480817 EDW50087.1 CM000363 CM002912 EDX08836.1 KMY96903.1 AE014296 AY069059 AAF47539.2 AAL39204.1 AHN57927.1 KQ971342 KYB27569.1 GEZM01101393 JAV52667.1 KQ979347 KYN21874.1 CCAG010023908 CCAG010023909 JXJN01010337 ADMH02001806 ETN61056.1 JTDY01000159 KOB78531.1 ACPB03017241 KK853069 KDR11826.1 GBBI01000582 JAC18130.1 DS235882 EEB20049.1 CH933816 EDW17249.1 GAHY01000041 JAA77469.1 CP012525 ALC44399.1 KZ288432 PBC25812.1 GBHO01018245 JAG25359.1 JR040074 AEY58968.1 KQ977304 KYN03801.1 GDHC01020240 JAP98388.1 CH940647 EDW68716.1 KQ761783 OAD57057.1 KQ434831 KZC07812.1 KQ435989 KOX67715.1 GL450772 EFN80505.1 KQ976394 KYM93169.1 ADTU01008269 ADTU01008270 KK107119 QOIP01000009 EZA58734.1 RLU18380.1 KQ414654 KOC65891.1 GL439967 EFN66483.1 KQ981880 KYN33937.1 CH916366 EDV95899.1 GDAI01002597 JAI15006.1 GECU01016585 GECU01012688 JAS91121.1 JAS95018.1 GECZ01021441 GECZ01019659 JAS48328.1 JAS50110.1 LJIG01000521 KRT86419.1 GEDC01005352 JAS31946.1 EFA03684.2 AJVK01008234 AJWK01005810 AJWK01005811 AJWK01005812 AJWK01005813 AXCM01022306 GFXV01005709 MBW17514.1 ABLF02038860 AK340708 BAH71266.1 LJIJ01000801 ODM94485.1 IACF01005413 LAB70998.1 KK855041 PTY20472.1
ODYU01004946 SOQ45337.1 KZ150103 PZC73465.1 BABH01037020 GAPW01005393 JAC08205.1 GFDL01002057 JAV32988.1 UFQS01000127 UFQT01000127 SSX00161.1 SSX20541.1 AAAB01008807 EAA04565.5 AXCN02000377 GAMD01000697 JAB00894.1 CH477572 EAT38864.1 JRES01000810 KNC28201.1 GAKP01014844 JAC44108.1 GBYB01000761 JAG70528.1 GDHF01028906 GDHF01022416 GDHF01011113 JAI23408.1 JAI29898.1 JAI41201.1 CH964168 EDW80151.1 GFDF01010914 JAV03170.1 CH902618 EDV38792.1 CM000159 EDW92875.1 GBXI01004199 JAD10093.1 CH954178 EDV50200.1 APGK01027575 APGK01027576 APGK01027577 APGK01027578 APGK01027579 KB740664 KB631791 ENN79682.1 ERL86099.1 ATLV01024587 KE525352 KFB51719.1 GL763924 EFZ19021.1 APCN01000205 CH479206 EDW30903.1 CH379069 EAL29459.2 KRT08027.1 OUUW01000002 SPP76966.1 CH480817 EDW50087.1 CM000363 CM002912 EDX08836.1 KMY96903.1 AE014296 AY069059 AAF47539.2 AAL39204.1 AHN57927.1 KQ971342 KYB27569.1 GEZM01101393 JAV52667.1 KQ979347 KYN21874.1 CCAG010023908 CCAG010023909 JXJN01010337 ADMH02001806 ETN61056.1 JTDY01000159 KOB78531.1 ACPB03017241 KK853069 KDR11826.1 GBBI01000582 JAC18130.1 DS235882 EEB20049.1 CH933816 EDW17249.1 GAHY01000041 JAA77469.1 CP012525 ALC44399.1 KZ288432 PBC25812.1 GBHO01018245 JAG25359.1 JR040074 AEY58968.1 KQ977304 KYN03801.1 GDHC01020240 JAP98388.1 CH940647 EDW68716.1 KQ761783 OAD57057.1 KQ434831 KZC07812.1 KQ435989 KOX67715.1 GL450772 EFN80505.1 KQ976394 KYM93169.1 ADTU01008269 ADTU01008270 KK107119 QOIP01000009 EZA58734.1 RLU18380.1 KQ414654 KOC65891.1 GL439967 EFN66483.1 KQ981880 KYN33937.1 CH916366 EDV95899.1 GDAI01002597 JAI15006.1 GECU01016585 GECU01012688 JAS91121.1 JAS95018.1 GECZ01021441 GECZ01019659 JAS48328.1 JAS50110.1 LJIG01000521 KRT86419.1 GEDC01005352 JAS31946.1 EFA03684.2 AJVK01008234 AJWK01005810 AJWK01005811 AJWK01005812 AJWK01005813 AXCM01022306 GFXV01005709 MBW17514.1 ABLF02038860 AK340708 BAH71266.1 LJIJ01000801 ODM94485.1 IACF01005413 LAB70998.1 KK855041 PTY20472.1
Proteomes
UP000053268
UP000007151
UP000053240
UP000005204
UP000075900
UP000075920
+ More
UP000007062 UP000075882 UP000075886 UP000075880 UP000076408 UP000008820 UP000037069 UP000075884 UP000007798 UP000095300 UP000007801 UP000002282 UP000075902 UP000008711 UP000019118 UP000030742 UP000030765 UP000192221 UP000075903 UP000075881 UP000075885 UP000075840 UP000008744 UP000001819 UP000076407 UP000268350 UP000001292 UP000000304 UP000000803 UP000007266 UP000095301 UP000078492 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000000673 UP000069272 UP000037510 UP000015103 UP000027135 UP000009046 UP000009192 UP000092553 UP000242457 UP000005203 UP000078542 UP000008792 UP000076502 UP000053105 UP000008237 UP000078540 UP000005205 UP000053097 UP000279307 UP000053825 UP000000311 UP000078541 UP000001070 UP000091820 UP000002358 UP000092462 UP000092461 UP000075883 UP000007819 UP000094527
UP000007062 UP000075882 UP000075886 UP000075880 UP000076408 UP000008820 UP000037069 UP000075884 UP000007798 UP000095300 UP000007801 UP000002282 UP000075902 UP000008711 UP000019118 UP000030742 UP000030765 UP000192221 UP000075903 UP000075881 UP000075885 UP000075840 UP000008744 UP000001819 UP000076407 UP000268350 UP000001292 UP000000304 UP000000803 UP000007266 UP000095301 UP000078492 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000000673 UP000069272 UP000037510 UP000015103 UP000027135 UP000009046 UP000009192 UP000092553 UP000242457 UP000005203 UP000078542 UP000008792 UP000076502 UP000053105 UP000008237 UP000078540 UP000005205 UP000053097 UP000279307 UP000053825 UP000000311 UP000078541 UP000001070 UP000091820 UP000002358 UP000092462 UP000092461 UP000075883 UP000007819 UP000094527
Interpro
IPR029693
TMEM35A
+ More
IPR040399 TM35A/B
IPR032808 DoxX
IPR019180 Oxidoreductase-like_N
IPR008333 Cbr1-like_FAD-bd_dom
IPR039261 FNR_nucleotide-bd
IPR001834 CBR-like
IPR017938 Riboflavin_synthase-like_b-brl
IPR001433 OxRdtase_FAD/NAD-bd
IPR017927 FAD-bd_FR_type
IPR001709 Flavoprot_Pyr_Nucl_cyt_Rdtase
IPR040399 TM35A/B
IPR032808 DoxX
IPR019180 Oxidoreductase-like_N
IPR008333 Cbr1-like_FAD-bd_dom
IPR039261 FNR_nucleotide-bd
IPR001834 CBR-like
IPR017938 Riboflavin_synthase-like_b-brl
IPR001433 OxRdtase_FAD/NAD-bd
IPR017927 FAD-bd_FR_type
IPR001709 Flavoprot_Pyr_Nucl_cyt_Rdtase
Gene 3D
ProteinModelPortal
A0A194PXE3
A0A212F0N4
A0A194R0N4
A0A2H1VWW1
A0A2W1BID4
H9J5B2
+ More
A0A023EH56 A0A182RSQ0 A0A1Q3FZK6 A0A182WJ68 A0A336LU73 Q7PT42 A0A182KRD5 A0A182Q2J9 A0A182ITK3 A0A182XYX9 T1EA69 Q16W86 A0A0L0C7H7 A0A034VRC8 A0A0C9R1P8 A0A0K8UA00 A0A182NHT8 B4N712 A0A1L8DA13 A0A1I8PR35 B3M7R9 B4PDR0 A0A0A1XHW1 A0A182UFU8 B3NES7 N6ULU9 A0A084WNC5 A0A1W4VV60 E9IK90 A0A182UZY9 A0A182KES5 A0A182PQD5 A0A182I3C5 B4H3G3 Q2LZP8 A0A182X3P4 A0A3B0JUD0 B4HVX2 B4QMH9 Q8T0T9 A0A139WI43 A0A1Y1K1H0 A0A1I8MBB6 A0A151JA11 A0A1B0FM45 A0A1A9Y9M3 A0A1A9VYK9 A0A1B0B9A0 A0A1A9Z3D5 W5JAN0 A0A182FF60 A0A0L7LTG0 T1I5P7 A0A067QQZ6 A0A023F9R2 E0W343 B4L974 R4G5E4 A0A0M3QWM3 A0A2A3E3D9 A0A0A9XZL1 A0A088A6Z7 V9ICU9 A0A195CSX8 A0A146KQV3 B4LBT9 A0A310SBE3 A0A154P7G6 A0A0M8ZNB0 E2BUW8 A0A195BX10 A0A158P3M8 A0A026WUE9 A0A0L7R561 E2AJ43 A0A195F192 B4J1X4 A0A1A9X2W2 A0A0K8TLH2 A0A1B6IW00 A0A1B6FDR1 A0A0T6BGG1 K7ITS8 A0A1B6E1Y9 D6WJE9 A0A1B0DPS7 A0A1B0CBS9 A0A182M9S9 A0A2H8TTE1 J9K3Z3 C4WTP6 A0A1D2MNR0 A0A2P2IAE9 A0A2R7WLI4
A0A023EH56 A0A182RSQ0 A0A1Q3FZK6 A0A182WJ68 A0A336LU73 Q7PT42 A0A182KRD5 A0A182Q2J9 A0A182ITK3 A0A182XYX9 T1EA69 Q16W86 A0A0L0C7H7 A0A034VRC8 A0A0C9R1P8 A0A0K8UA00 A0A182NHT8 B4N712 A0A1L8DA13 A0A1I8PR35 B3M7R9 B4PDR0 A0A0A1XHW1 A0A182UFU8 B3NES7 N6ULU9 A0A084WNC5 A0A1W4VV60 E9IK90 A0A182UZY9 A0A182KES5 A0A182PQD5 A0A182I3C5 B4H3G3 Q2LZP8 A0A182X3P4 A0A3B0JUD0 B4HVX2 B4QMH9 Q8T0T9 A0A139WI43 A0A1Y1K1H0 A0A1I8MBB6 A0A151JA11 A0A1B0FM45 A0A1A9Y9M3 A0A1A9VYK9 A0A1B0B9A0 A0A1A9Z3D5 W5JAN0 A0A182FF60 A0A0L7LTG0 T1I5P7 A0A067QQZ6 A0A023F9R2 E0W343 B4L974 R4G5E4 A0A0M3QWM3 A0A2A3E3D9 A0A0A9XZL1 A0A088A6Z7 V9ICU9 A0A195CSX8 A0A146KQV3 B4LBT9 A0A310SBE3 A0A154P7G6 A0A0M8ZNB0 E2BUW8 A0A195BX10 A0A158P3M8 A0A026WUE9 A0A0L7R561 E2AJ43 A0A195F192 B4J1X4 A0A1A9X2W2 A0A0K8TLH2 A0A1B6IW00 A0A1B6FDR1 A0A0T6BGG1 K7ITS8 A0A1B6E1Y9 D6WJE9 A0A1B0DPS7 A0A1B0CBS9 A0A182M9S9 A0A2H8TTE1 J9K3Z3 C4WTP6 A0A1D2MNR0 A0A2P2IAE9 A0A2R7WLI4
Ontologies
PANTHER
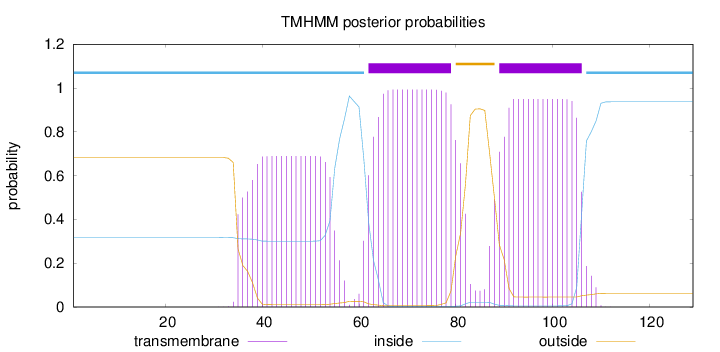
Topology
Length:
129
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
50.53488
Exp number, first 60 AAs:
13.70809
Total prob of N-in:
0.31712
POSSIBLE N-term signal
sequence
inside
1 - 61
TMhelix
62 - 79
outside
80 - 88
TMhelix
89 - 106
inside
107 - 129
Population Genetic Test Statistics
Pi
55.881718
Theta
18.501586
Tajima's D
-1.642877
CLR
4.688937
CSRT
0.0425478726063697
Interpretation
Uncertain
